Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
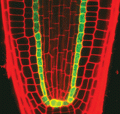
2329: Planting roots
2329: Planting roots
At the root tips of the mustard plant Arabidopsis thaliana (red), two proteins work together to control the uptake of water and nutrients. When the cell division-promoting protein called Short-root moves from the center of the tip outward, it triggers the production of another protein (green) that confines Short-root to the nutrient-filtering endodermis. The mechanism sheds light on how genes and proteins interact in a model organism and also could inform the engineering of plants.
Philip Benfey, Duke University
View Media
3627: Larvae from the parasitic worm that causes schistosomiasis
3627: Larvae from the parasitic worm that causes schistosomiasis
The parasitic worm that causes schistosomiasis hatches in water and grows up in a freshwater snail, as shown here. Once mature, the worm swims back into the water, where it can infect people through skin contact. Initially, an infected person might have a rash, itchy skin, or flu-like symptoms, but the real damage is done over time to internal organs.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bo Wang and Phillip A. Newmark, University of Illinois at Urbana-Champaign, 2013 FASEB BioArt winner
View Media
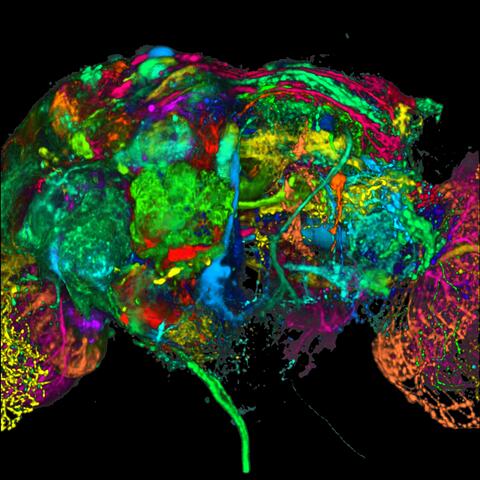
5868: Color coding of the Drosophila brain - black background
5868: Color coding of the Drosophila brain - black background
This image results from a research project to visualize which regions of the adult fruit fly (Drosophila) brain derive from each neural stem cell. First, researchers collected several thousand fruit fly larvae and fluorescently stained a random stem cell in the brain of each. The idea was to create a population of larvae in which each of the 100 or so neural stem cells was labeled at least once. When the larvae grew to adults, the researchers examined the flies’ brains using confocal microscopy. With this technique, the part of a fly’s brain that derived from a single, labeled stem cell “lights up.” The scientists photographed each brain and digitally colorized its lit-up area. By combining thousands of such photos, they created a three-dimensional, color-coded map that shows which part of the Drosophila brain comes from each of its ~100 neural stem cells. In other words, each colored region shows which neurons are the progeny or “clones” of a single stem cell. This work established a hierarchical structure as well as nomenclature for the neurons in the Drosophila brain. Further research will relate functions to structures of the brain.
Related to image 5838 and video 5843.
Related to image 5838 and video 5843.
Yong Wan from Charles Hansen’s lab, University of Utah. Data preparation and visualization by Masayoshi Ito in the lab of Kei Ito, University of Tokyo.
View Media
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
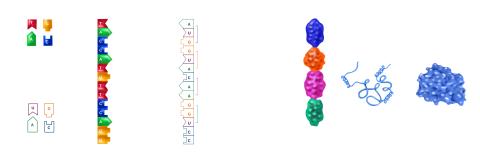
2509: From DNA to Protein
2509: From DNA to Protein
Nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
See image 2510 for a labeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media
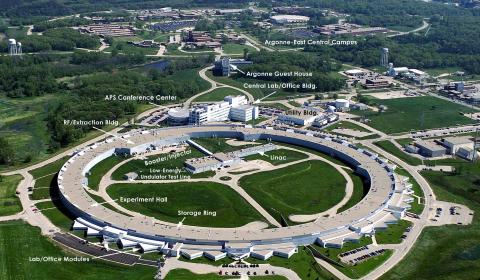
2358: Advanced Photon Source (APS) at Argonne National Lab
2358: Advanced Photon Source (APS) at Argonne National Lab
The intense X-rays produced by synchrotrons such as the Advanced Photon Source are ideally suited for protein structure determination. Using synchrotron X-rays and advanced computers scientists can determine protein structures at a pace unheard of decades ago.
Southeast Collaboratory for Structural Genomics
View Media
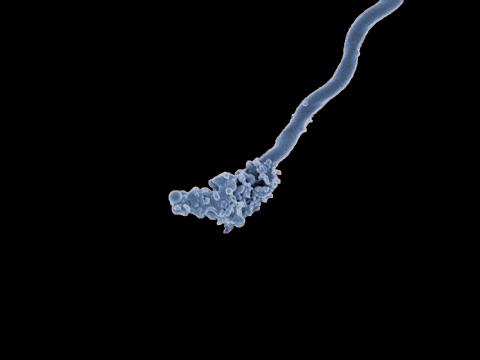
3585: Relapsing fever bacterium (gray) and red blood cells
3585: Relapsing fever bacterium (gray) and red blood cells
Relapsing fever is caused by a bacterium and transmitted by certain soft-bodied ticks or body lice. The disease is seldom fatal in humans, but it can be very serious and prolonged. This scanning electron micrograph shows Borrelia hermsii (green), one of the bacterial species that causes the disease, interacting with red blood cells. Micrograph by Robert Fischer, NIAID. Related to image 3586.
For more information about relapsing fever, see https://www.cdc.gov/relapsing-fever/index.html.
This image is part of the Life: Magnified collection, which was displayed in the Gateway Gallery at Washington Dulles International Airport June 3, 2014, to January 21, 2015.
For more information about relapsing fever, see https://www.cdc.gov/relapsing-fever/index.html.
This image is part of the Life: Magnified collection, which was displayed in the Gateway Gallery at Washington Dulles International Airport June 3, 2014, to January 21, 2015.
NIAID
View Media
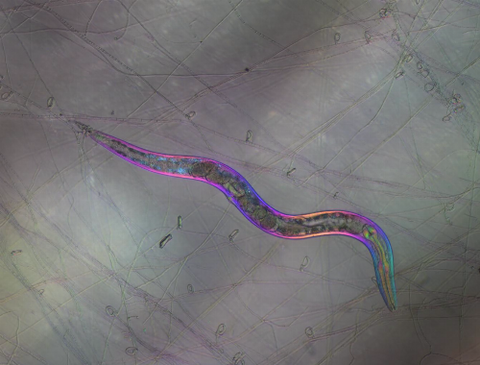
6963: C. elegans trapped by carnivorous fungus
6963: C. elegans trapped by carnivorous fungus
Real-time footage of Caenorhabditis elegans, a tiny roundworm, trapped by a carnivorous fungus, Arthrobotrys dactyloides. This fungus makes ring traps in response to the presence of C. elegans. When a worm enters a ring, the trap rapidly constricts so that the worm cannot move away, and the fungus then consumes the worm. The size of the imaged area is 0.7mm x 0.9mm.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
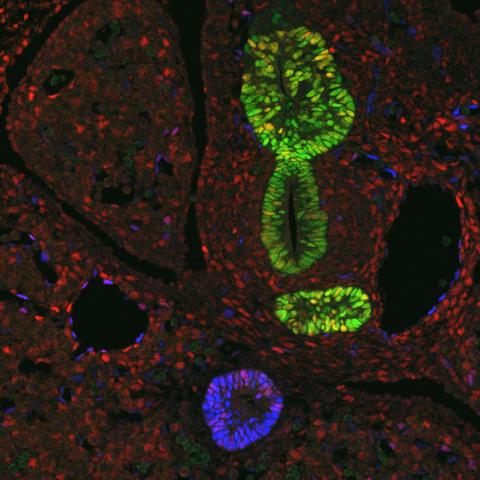
3440: Transcription factor Sox17 controls embryonic development of certain internal organs
3440: Transcription factor Sox17 controls embryonic development of certain internal organs
During embryonic development, transcription factors (proteins that regulate gene expression) govern the differentiation of cells into separate tissues and organs. Researchers at Cincinnati Children's Hospital Medical Center used mice to study the development of certain internal organs, including the liver, pancreas, duodenum (beginning part of the small intestine), gall bladder and bile ducts. They discovered that transcription factor Sox17 guides some cells to develop into liver cells and others to become part of the pancreas or biliary system (gall bladder, bile ducts and associated structures). The separation of these two distinct cell types (liver versus pancreas/biliary system) is complete by embryonic day 8.5 in mice. The transcription factors PDX1 and Hes1 are also known to be involved in embryonic development of the pancreas and biliary system. This image shows mouse cells at embryonic day 10.5. The green areas show cells that will develop into the pancreas and/or duodenum(PDX1 is labeled green). The blue area near the bottom will become the gall bladder and the connecting tubes (common duct and cystic duct) that attach the gall bladder to the liver and pancreas (Sox17 is labeled blue). The transcription factor Hes1 is labeled red. The image was not published. A similar image (different plane of the section) was published in: Sox17 Regulates Organ Lineage Segregation of Ventral Foregut Progenitor Cells Jason R. Spence, Alex W. Lange, Suh-Chin J. Lin, Klaus H. Kaestner, Andrew M. Lowy, Injune Kim, Jeffrey A. Whitsett and James M. Wells, Developmental Cell, Volume 17, Issue 1, 62-74, 21 July 2009. doi:10.1016/j.devcel.2009.05.012
James M. Wells, Cincinnati Children's Hospital Medical Center
View Media

2414: Pig trypsin (3)
2414: Pig trypsin (3)
Crystals of porcine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
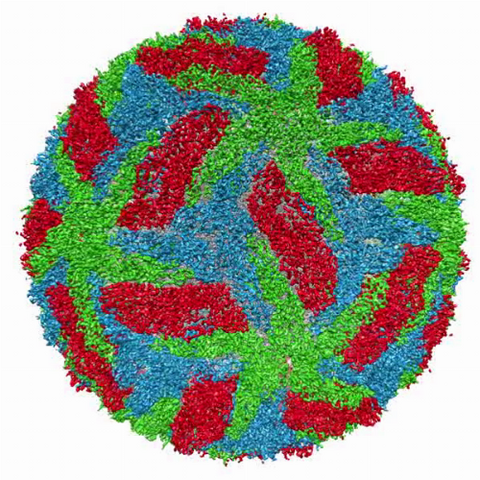
3748: Cryo-electron microscopy of the dengue virus showing protective membrane and membrane proteins
3748: Cryo-electron microscopy of the dengue virus showing protective membrane and membrane proteins
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. Related to image 3756.
Hong Zhou, UCLA
View Media
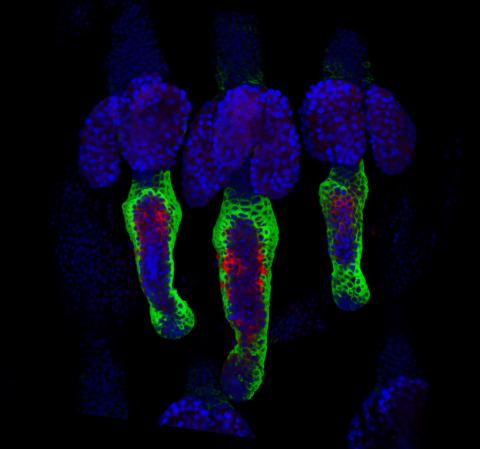
3497: Wound healing in process
3497: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3498 and 3500.
Related to images 3498 and 3500.
Hermann Steller, Rockefeller University
View Media
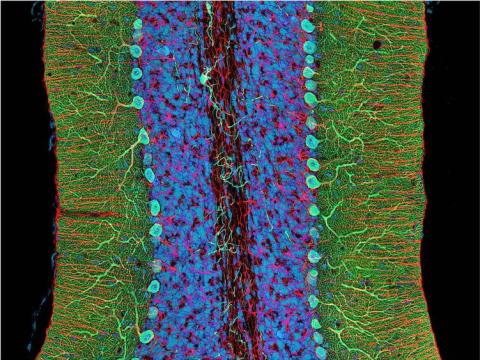
3371: Mouse cerebellum close-up
3371: Mouse cerebellum close-up
The cerebellum is the brain's locomotion control center. Every time you shoot a basketball, tie your shoe or chop an onion, your cerebellum fires into action. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills. For a lower magnification, see image 3639.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
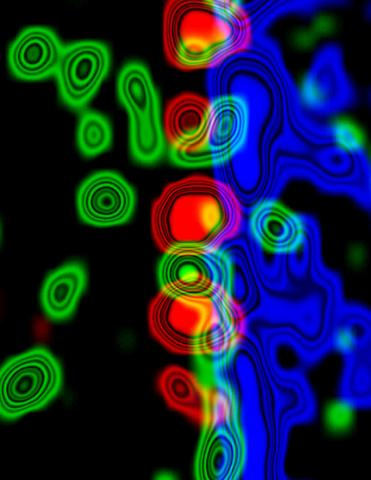
3734: Molecular interactions at the astrocyte nuclear membrane
3734: Molecular interactions at the astrocyte nuclear membrane
These ripples of color represent the outer membrane of the nucleus inside an astrocyte, a star-shaped cell inside the brain. Some proteins (green) act as keys to unlock other proteins (red) that form gates to let small molecules in and out of the nucleus (blue). Visualizing these different cell components at the boundary of the astrocyte nucleus enables researchers to study the molecular and physiological basis of neurological disorders, such as hydrocephalus, a condition in which too much fluid accumulates in the brain, and scar formation in brain tissue leading to abnormal neuronal activity affecting learning and memory. Scientists have now identified a pathway may be common to many of these brain diseases and begun to further examine it to find ways to treat certain brain diseases and injuries.
Katerina Akassoglou, Gladstone Institute for Neurological Disease & UCSF
View Media
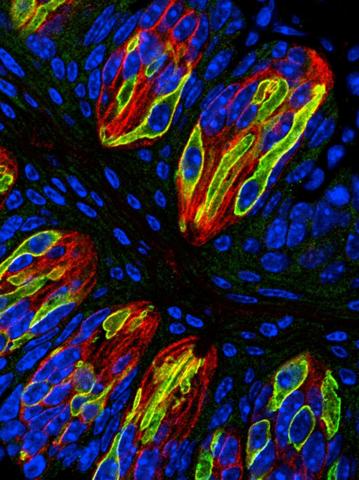
3444: Taste buds signal different tastes through ATP release
3444: Taste buds signal different tastes through ATP release
Taste buds in a mouse tongue epithelium with types I, II, and III taste cells visualized by cell-type-specific fluorescent antibodies. Type II taste bud cells signal sweet, bitter, and umami tastes to the central nervous system by releasing ATP through the voltage-gated ion channel CALHM1. Researchers used a confocal microscope to capture this image, which shows all taste buds in red, type II taste buds in green, and DNA in blue.
More information about this work can be found in the Nature letter "CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes” by Taruno et. al.
More information about this work can be found in the Nature letter "CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes” by Taruno et. al.
Aki Taruno, Perelman School of Medicine, University of Pennsylvania
View Media
6775: Tracking embryonic zebrafish cells
6775: Tracking embryonic zebrafish cells
To better understand cell movements in developing embryos, researchers isolated cells from early zebrafish embryos and grew them as clusters. Provided with the right signals, the clusters replicated some cell movements seen in intact embryos. Each line in this image depicts the movement of a single cell. The image was created using time-lapse confocal microscopy. Related to video 6776.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media

6751: Petri dish containing C. elegans
6751: Petri dish containing C. elegans
This Petri dish contains microscopic roundworms called Caenorhabditis elegans. Researchers used these particular worms to study how C. elegans senses the color of light in its environment.
H. Robert Horvitz and Dipon Ghosh, Massachusetts Institute of Technology.
View Media
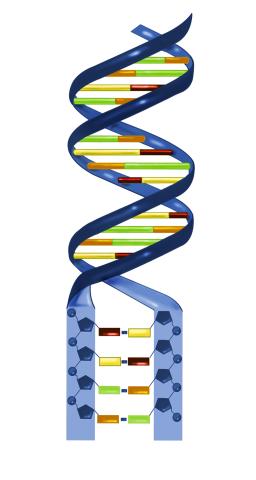
2541: Nucleotides make up DNA
2541: Nucleotides make up DNA
DNA consists of two long, twisted chains made up of nucleotides. Each nucleotide contains one base, one phosphate molecule, and the sugar molecule deoxyribose. The bases in DNA nucleotides are adenine, thymine, cytosine, and guanine. See image 2542 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
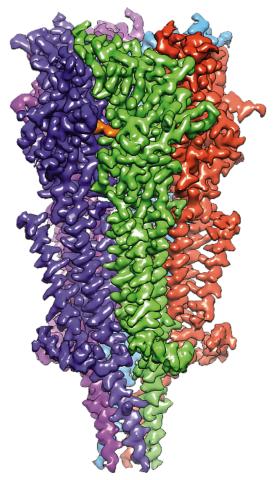
6579: Full-length serotonin receptor (ion channel)
6579: Full-length serotonin receptor (ion channel)
A 3D reconstruction, created using cryo-electron microscopy, of an ion channel known as the full-length serotonin receptor in complex with the antinausea drug granisetron (orange). Ion channels are proteins in cell membranes that help regulate many processes.
Sudha Chakrapani, Case Western Reserve University School of Medicine.
View Media
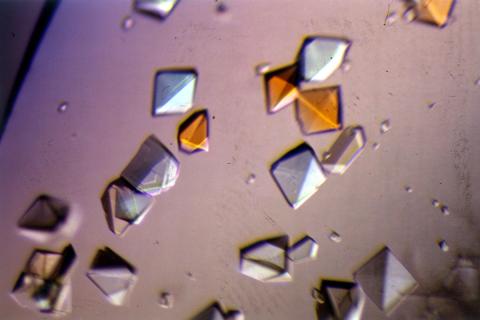
2412: Pig alpha amylase
2412: Pig alpha amylase
Crystals of porcine alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
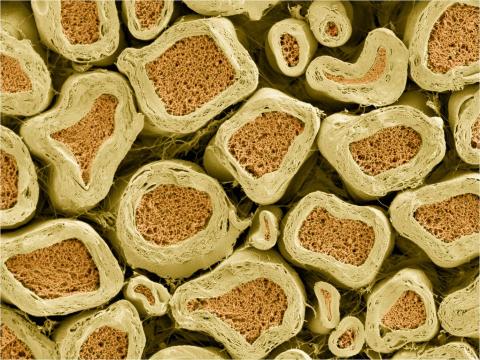
3397: Myelinated axons 2
3397: Myelinated axons 2
Top view of myelinated axons in a rat spinal root. Myelin is a type of fat that forms a sheath around and thus insulates the axon to protect it from losing the electrical current needed to transmit signals along the axon. The axoplasm inside the axon is shown in pink. Related to 3396.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

1069: Lab mice
1069: Lab mice
Many researchers use the mouse (Mus musculus) as a model organism to study mammalian biology. Mice carry out practically all the same life processes as humans and, because of their small size and short generation times, are easily raised in labs. Scientists studying a certain cellular activity or disease can choose from tens of thousands of specially bred strains of mice to select those prone to developing certain tumors, neurological diseases, metabolic disorders, premature aging, or other conditions.
Bill Branson, National Institutes of Health
View Media
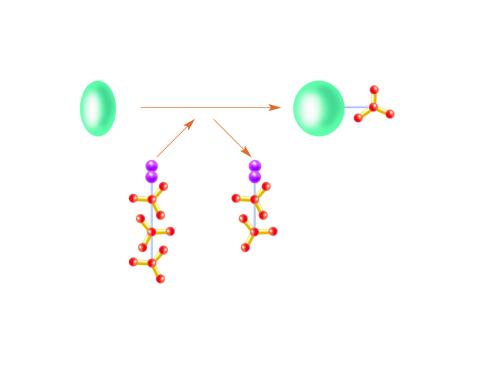
2534: Kinases
2534: Kinases
Kinases are enzymes that add phosphate groups (red-yellow structures) to proteins (green), assigning the proteins a code. In this reaction, an intermediate molecule called ATP (adenosine triphosphate) donates a phosphate group from itself, becoming ADP (adenosine diphosphate). See image 2535 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
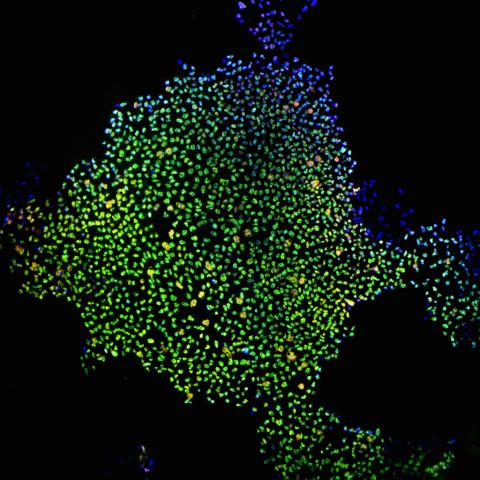
2604: Induced stem cells from adult skin 02
2604: Induced stem cells from adult skin 02
These cells are induced stem cells made from human adult skin cells that were genetically reprogrammed to mimic embryonic stem cells. The induced stem cells were made potentially safer by removing the introduced genes and the viral vector used to ferry genes into the cells, a loop of DNA called a plasmid. The work was accomplished by geneticist Junying Yu in the laboratory of James Thomson, a University of Wisconsin-Madison School of Medicine and Public Health professor and the director of regenerative biology for the Morgridge Institute for Research.
James Thomson, University of Wisconsin-Madison
View Media
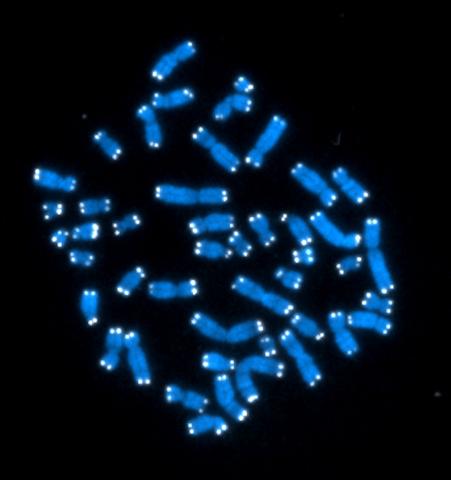
2626: Telomeres
2626: Telomeres
The 46 human chromosomes are shown in blue, with the telomeres appearing as white pinpoints. The DNA has already been copied, so each chromosome is actually made up of two identical lengths of DNA, each with its own two telomeres.
Hesed Padilla-Nash and Thomas Ried, the National Cancer Institute, a part of NIH
View Media
3597: DNA replication origin recognition complex (ORC)
3597: DNA replication origin recognition complex (ORC)
A study published in March 2012 used cryo-electron microscopy to determine the structure of the DNA replication origin recognition complex (ORC), a semi-circular, protein complex (yellow) that recognizes and binds DNA to start the replication process. The ORC appears to wrap around and bend approximately 70 base pairs of double stranded DNA (red and blue). Also shown is the protein Cdc6 (green), which is also involved in the initiation of DNA replication. Related to video 3307 that shows the structure from different angles. From a Brookhaven National Laboratory news release, "Study Reveals How Protein Machinery Binds and Wraps DNA to Start Replication."
Huilin Li, Brookhaven National Laboratory
View Media
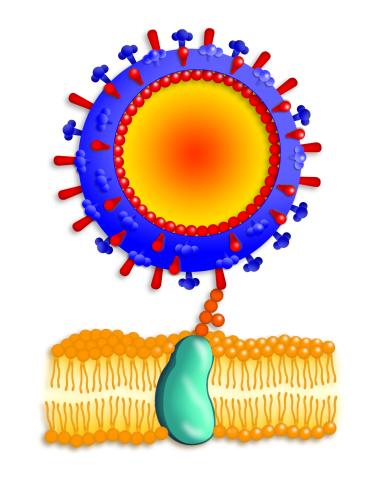
2425: Influenza virus attaches to host membrane
2425: Influenza virus attaches to host membrane
Influenza A infects a host cell when hemagglutinin grips onto glycans on its surface. Neuraminidase, an enzyme that chews sugars, helps newly made virus particles detach so they can infect other cells. Related to 213. Featured in the March 2006, issue of Findings in "Viral Voyages."
Crabtree + Company
View Media
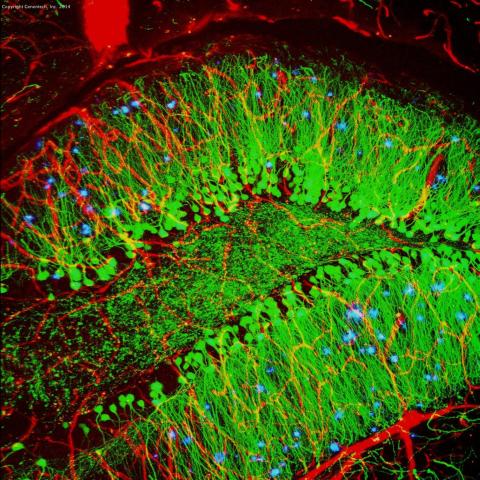
3604: Brain showing hallmarks of Alzheimer's disease
3604: Brain showing hallmarks of Alzheimer's disease
Along with blood vessels (red) and nerve cells (green), this mouse brain shows abnormal protein clumps known as plaques (blue). These plaques multiply in the brains of people with Alzheimer's disease and are associated with the memory impairment characteristic of the disease. Because mice have genomes nearly identical to our own, they are used to study both the genetic and environmental factors that trigger Alzheimer's disease. Experimental treatments are also tested in mice to identify the best potential therapies for human patients.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Alvin Gogineni, Genentech
View Media
1082: Natcher Building 02
1082: Natcher Building 02
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
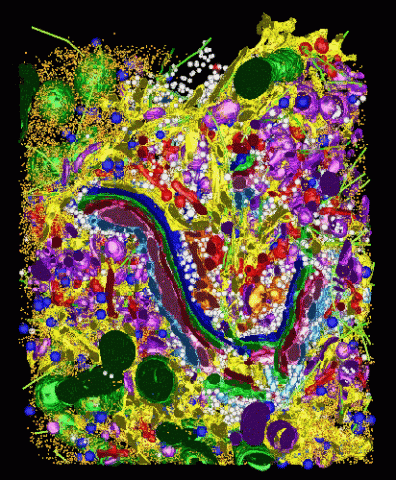
2308: Cellular metropolis
2308: Cellular metropolis
Like a major city, a cell teems with specialized workers that carry out its daily operations--making energy, moving proteins, or helping with other tasks. Researchers took microscopic pictures of thin layers of a cell and then combined them to make this 3-D image featuring color-coded organelles--the cell's "workers." Using this image, scientists can understand how these specialized components fit together in the cell's packed inner world.
Kathryn Howell, University of Colorado Health Sciences Center
View Media
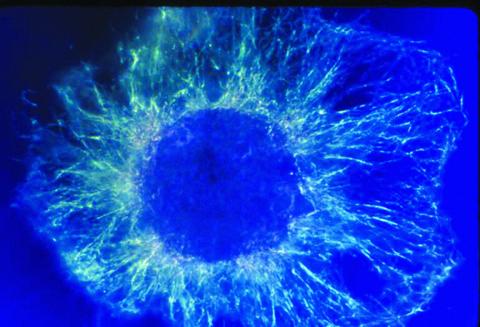
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media
2792: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 03
2792: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 03
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
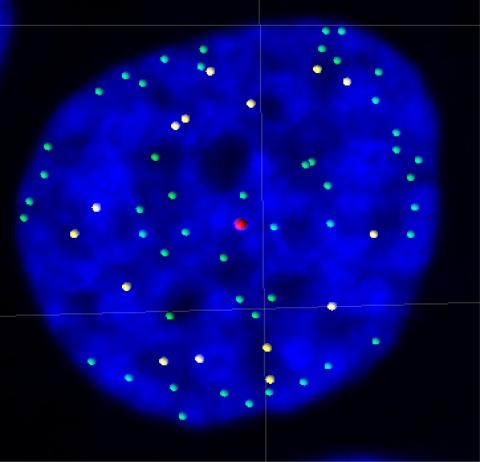
3484: Telomeres on outer edge of nucleus during cell division
3484: Telomeres on outer edge of nucleus during cell division
New research shows telomeres moving to the outer edge of the nucleus after cell division, suggesting these caps that protect chromosomes also may play a role in organizing DNA.
Laure Crabbe, Jamie Kasuboski and James Fitzpatrick, Salk Institute for Biological Studies
View Media
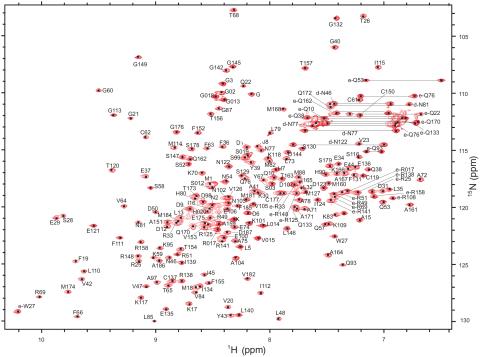
2299: 2-D NMR
2299: 2-D NMR
A two-dimensional NMR spectrum of a protein, in this case a 2D 1H-15N HSQC NMR spectrum of a 228 amino acid DNA/RNA-binding protein.
Dr. Xiaolian Gao's laboratory at the University of Houston
View Media
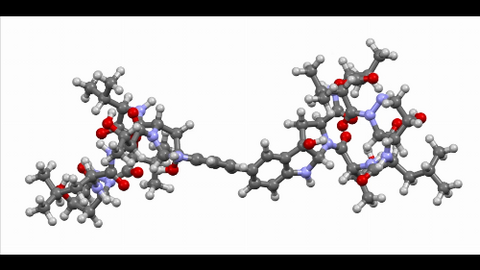
6851: Himastatin, 360-degree view
6851: Himastatin, 360-degree view
A 360-degree view of the molecule himastatin, which was first isolated from the bacterium Streptomyces himastatinicus. Himastatin shows antibiotic activity. The researchers who created this video developed a new, more concise way to synthesize himastatin so it can be studied more easily.
More information about the research that produced this video can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to images 6848 and 6850.
More information about the research that produced this video can be found in the Science paper “Total synthesis of himastatin” by D’Angelo et al.
Related to images 6848 and 6850.
Mohammad Movassaghi, Massachusetts Institute of Technology.
View Media

2376: Protein purification facility
2376: Protein purification facility
The Center for Eukaryotic Structural Genomics protein purification facility is responsible for purifying all recombinant proteins produced by the center. The facility performs several purification steps, monitors the quality of the processes, and stores information about the biochemical properties of the purified proteins in the facility database.
Center for Eukaryotic Structural Genomics
View Media

3424: White Poppy
3424: White Poppy
A white poppy. View cropped image of a poppy here 3423.
Judy Coyle, Donald Danforth Plant Science Center
View Media

2361: Chromium X-ray source
2361: Chromium X-ray source
In the determination of protein structures by X-ray crystallography, this unique soft (l = 2.29Å) X-ray source is used to collect anomalous scattering data from protein crystals containing light atoms such as sulfur, calcium, zinc and phosphorous. These data can be used to image the protein.
The Southeast Collaboratory for Structural Genomics
View Media
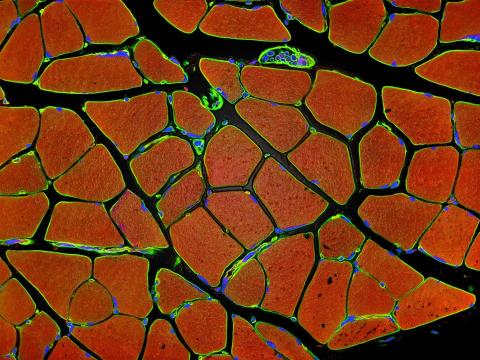
3677: Human skeletal muscle
3677: Human skeletal muscle
Cross section of human skeletal muscle. Image taken with a confocal fluorescent light microscope.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
6518: Biofilm formed by a pathogen
6518: Biofilm formed by a pathogen
A biofilm is a highly organized community of microorganisms that develops naturally on certain surfaces. These communities are common in natural environments and generally do not pose any danger to humans. Many microbes in biofilms have a positive impact on the planet and our societies. Biofilms can be helpful in treatment of wastewater, for example. This dime-sized biofilm, however, was formed by the opportunistic pathogen Pseudomonas aeruginosa. Under some conditions, this bacterium can infect wounds that are caused by severe burns. The bacterial cells release a variety of materials to form an extracellular matrix, which is stained red in this photograph. The matrix holds the biofilm together and protects the bacteria from antibiotics and the immune system.
Scott Chimileski, Ph.D., and Roberto Kolter, Ph.D., Harvard Medical School.
View Media
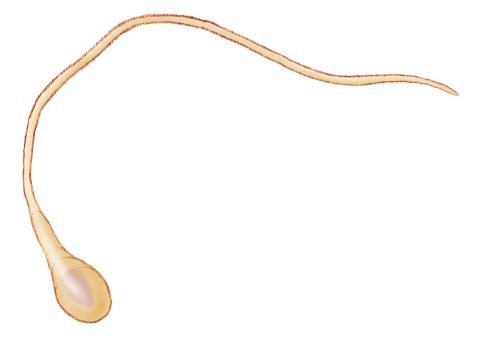
1293: Sperm cell
2566: Haplotypes
2566: Haplotypes
Haplotypes are combinations of gene variants that are likely to be inherited together within the same chromosomal region. In this example, an original haplotype (top) evolved over time to create three newer haplotypes that each differ by a few nucleotides (red). See image 2567 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
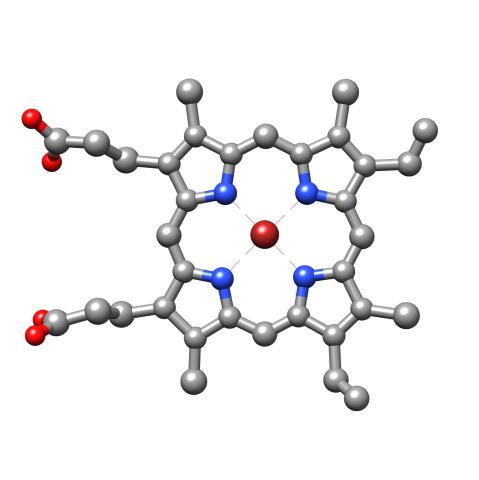
3539: Structure of heme, top view
3539: Structure of heme, top view
Molecular model of the struture of heme. Heme is a small, flat molecule with an iron ion (dark red) at its center. Heme is an essential component of hemoglobin, the protein in blood that carries oxygen throughout our bodies. This image first appeared in the September 2013 issue of Findings Magazine. View side view of heme here 3540.
Rachel Kramer Green, RCSB Protein Data Bank
View Media
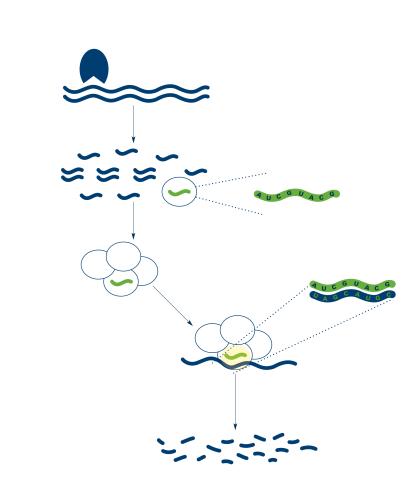
2558: RNA interference
2558: RNA interference
RNA interference or RNAi is a gene-silencing process in which double-stranded RNAs trigger the destruction of specific RNAs. See 2559 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media

1120: Superconducting magnet
1120: Superconducting magnet
Superconducting magnet for NMR research, from the February 2003 profile of Dorothee Kern in Findings.
Mike Lovett
View Media
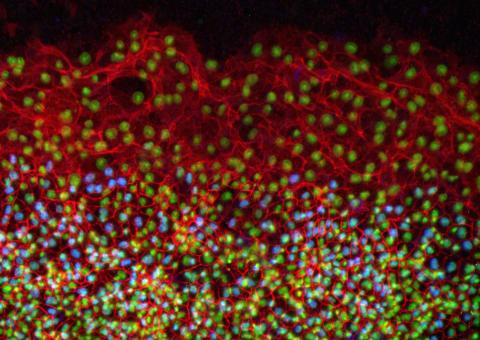
2808: Cell proliferation in a quail embryo
2808: Cell proliferation in a quail embryo
Image showing that the edge zone (top of image) of the quail embryo shows no proliferating cells (cyan), unlike the interior zone (bottom of image). Non-proliferating cell nuclei are labeled green. This image was obtained as part of a study to understand cell migration in embryos. More specifically, cell proliferation at the edge of the embryo was studied by examining the cellular uptake of a chemical compound called BrDU, which incorporates into the DNA during the S-phase of the cell cycle. Here, the cells that are positive for BrDU uptake are labeled in cyan, while other non-proliferating cell nuclei are labeled green. Notice that the vast majority of BrDU+ cells are located far away from the edge, indicating that edge cells are mostly non-proliferating. An NIGMS grant to Professor Garcia was used to purchase the confocal microscope that collected this image. Related to image 2807 and video 2809.
Andrés Garcia, Georgia Tech
View Media
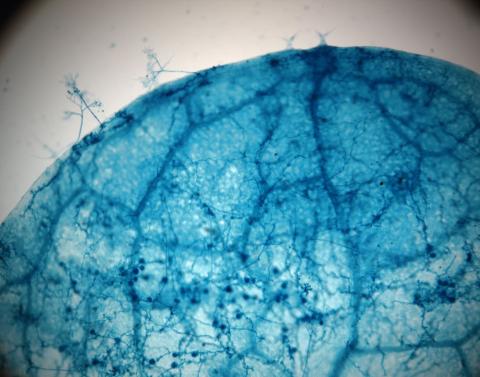
2782: Disease-susceptible Arabidopsis leaf
2782: Disease-susceptible Arabidopsis leaf
This is a magnified view of an Arabidopsis thaliana leaf after several days of infection with the pathogen Hyaloperonospora arabidopsidis. The pathogen's blue hyphae grow throughout the leaf. On the leaf's edges, stalk-like structures called sporangiophores are beginning to mature and will release the pathogen's spores. Inside the leaf, the large, deep blue spots are structures called oopsorangia, also full of spores. Compare this response to that shown in Image 2781. Jeff Dangl has been funded by NIGMS to study the interactions between pathogens and hosts that allow or suppress infection.
Jeff Dangl, University of North Carolina, Chapel Hill
View Media
2442: Hydra 06
2442: Hydra 06
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
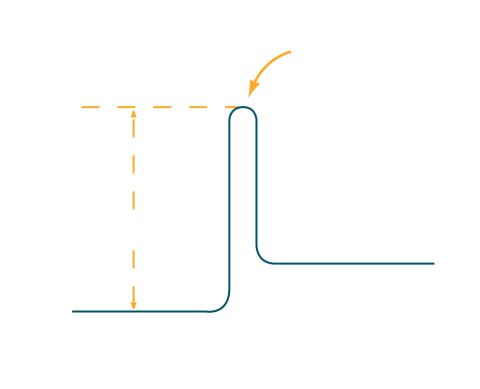
2525: Activation energy
2525: Activation energy
To become products, reactants must overcome an energy hill. See image 2526 for a labeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
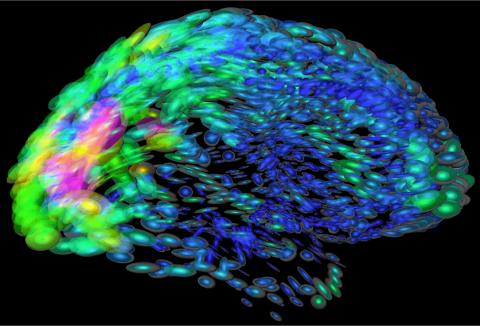
2419: Mapping brain differences
2419: Mapping brain differences
This image of the human brain uses colors and shapes to show neurological differences between two people. The blurred front portion of the brain, associated with complex thought, varies most between the individuals. The blue ovals mark areas of basic function that vary relatively little. Visualizations like this one are part of a project to map complex and dynamic information about the human brain, including genes, enzymes, disease states, and anatomy. The brain maps represent collaborations between neuroscientists and experts in math, statistics, computer science, bioinformatics, imaging, and nanotechnology.
Arthur Toga, University of California, Los Angeles
View Media
