Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
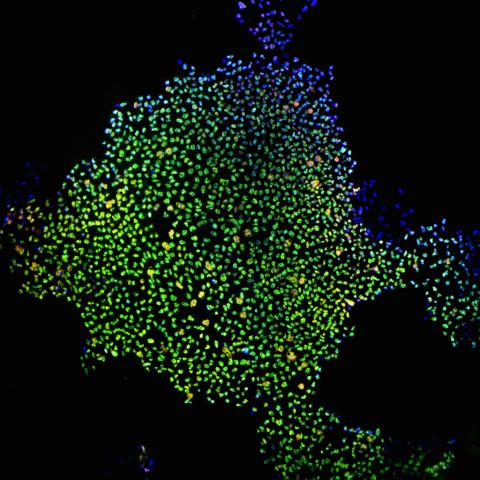
2604: Induced stem cells from adult skin 02
2604: Induced stem cells from adult skin 02
These cells are induced stem cells made from human adult skin cells that were genetically reprogrammed to mimic embryonic stem cells. The induced stem cells were made potentially safer by removing the introduced genes and the viral vector used to ferry genes into the cells, a loop of DNA called a plasmid. The work was accomplished by geneticist Junying Yu in the laboratory of James Thomson, a University of Wisconsin-Madison School of Medicine and Public Health professor and the director of regenerative biology for the Morgridge Institute for Research.
James Thomson, University of Wisconsin-Madison
View Media

2411: Fungal lipase (2)
2411: Fungal lipase (2)
Crystals of fungal lipase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
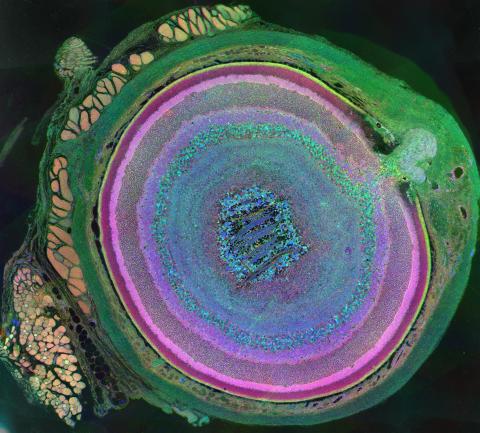
3641: A mammalian eye has approximately 70 different cell types
3641: A mammalian eye has approximately 70 different cell types
The incredible complexity of a mammalian eye (in this case from a mouse) is captured here. Each color represents a different type of cell. In total, there are nearly 70 different cell types, including the retina's many rings and the peach-colored muscle cells clustered on the left.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bryan William Jones and Robert E. Marc, University of Utah
View Media
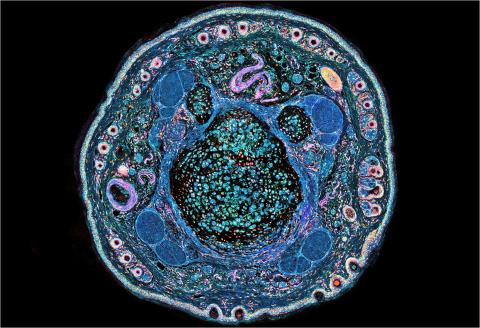
3395: NCMIR mouse tail
3395: NCMIR mouse tail
Stained cross section of a mouse tail.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
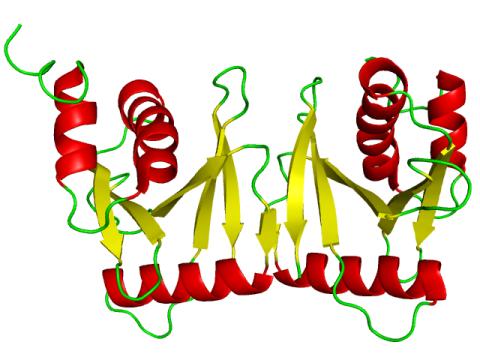
2351: tRNA splicing enzyme endonuclease in humans
2351: tRNA splicing enzyme endonuclease in humans
An NMR solution structure model of the transfer RNA splicing enzyme endonuclease in humans (subunit Sen15). This represents the first structure of a eukaryotic tRNA splicing endonuclease subunit.
Center for Eukaryotic Structural Genomics, PSI
View Media
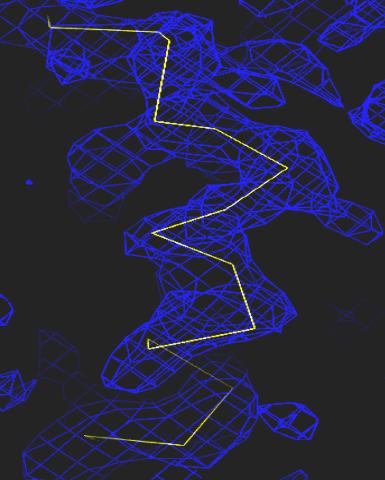
2354: Section of an electron density map
2354: Section of an electron density map
Electron density maps such as this one are generated from the diffraction patterns of X-rays passing through protein crystals. These maps are then used to generate a model of the protein's structure by fitting the protein's amino acid sequence (yellow) into the observed electron density (blue).
The Southeast Collaboratory for Structural Genomics
View Media
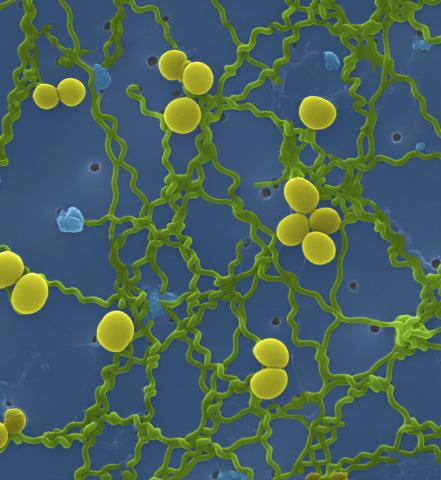
1166: Leptospira bacteria
1166: Leptospira bacteria
Leptospira, shown here in green, is a type (genus) of elongated, spiral-shaped bacteria. Infection can cause Weil's disease, a kind of jaundice, in humans.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
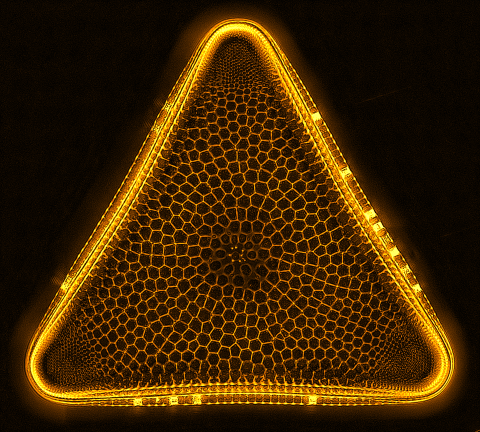
6962: Trigonium diatom
6962: Trigonium diatom
A Trigonium diatom imaged by a quantitative orientation-independent differential interference contrast (OI-DIC) microscope. Diatoms are single-celled photosynthetic algae with mineralized cell walls that contain silica and provide protection and support. These organisms form an important part of the plankton at the base of the marine and freshwater food chains. The width of this image is 90 μm.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
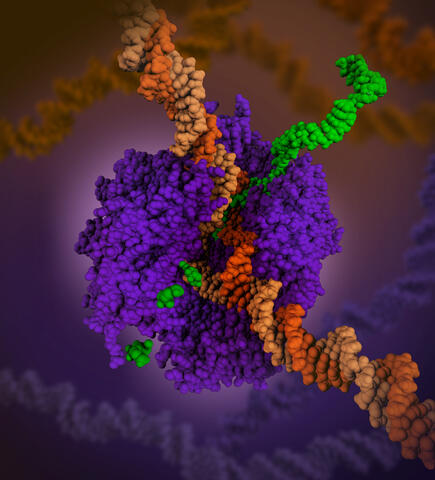
6993: RNA polymerase
6993: RNA polymerase
RNA polymerase (purple) is a complex enzyme at the heart of transcription. During this process, the enzyme unwinds the DNA double helix and uses one strand (darker orange) as a template to create the single-stranded messenger RNA (green), later used by ribosomes for protein synthesis.
From the RNA polymerase II elongation complex of Saccharomyces cerevisiae (PDB entry 1I6H) as seen in PDB-101's What is a Protein? video.
From the RNA polymerase II elongation complex of Saccharomyces cerevisiae (PDB entry 1I6H) as seen in PDB-101's What is a Protein? video.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
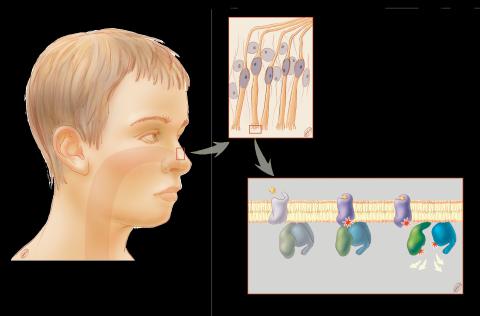
1291: Olfactory system
1291: Olfactory system
Sensory organs have cells equipped for detecting signals from the environment, such as odors. Receptors in the membranes of nerve cells in the nose bind to odor molecules, triggering a cascade of chemical reactions tranferred by G proteins into the cytoplasm.
Judith Stoffer
View Media
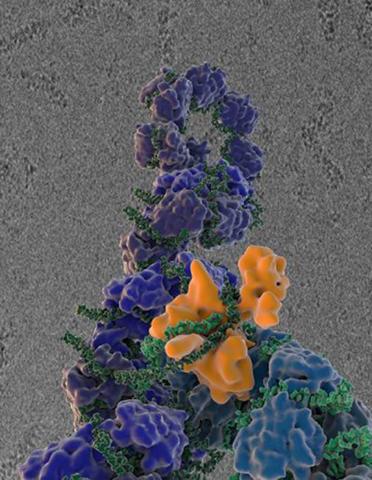
3434: Flu virus proteins during self-replication
3434: Flu virus proteins during self-replication
Influenza (flu) virus proteins in the act of self-replication. Viral nucleoprotein (blue) encapsidates [encapsulates] the RNA genome (green). The influenza virus polymerase (orange) reads and copies the RNA genome. In the background is an image of influenza virus ribonucleoprotein complexes observed using cryo-electron microscopy. This image is from a November 2012 News Release.
Scripps Research Institute in La Jolla, CA
View Media
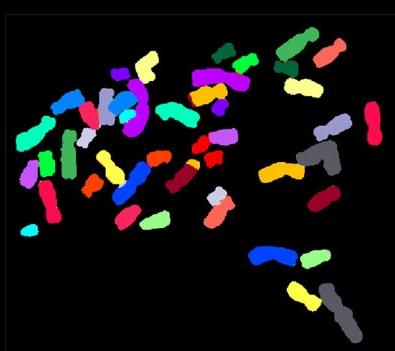
2312: Color-coded chromosomes
2312: Color-coded chromosomes
By mixing fluorescent dyes like an artist mixes paints, scientists are able to color code individual chromosomes. The technique, abbreviated multicolor-FISH, allows researchers to visualize genetic abnormalities often linked to disease. In this image, "painted" chromosomes from a person with a hereditary disease called Werner Syndrome show where a piece of one chromosome has fused to another (see the gold-tipped maroon chromosome in the center). As reported by molecular biologist Jan Karlseder of the Salk Institute for Biological Studies, such damage is typical among people with this rare syndrome.
Anna Jauch, Institute of Human Genetics, Heidelberg, Germany
View Media

6770: Group of Culex quinquefasciatus mosquito larvae
6770: Group of Culex quinquefasciatus mosquito larvae
Mosquito larvae with genes edited by CRISPR. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this image developed a gene-editing toolkit for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to image 6769 and video 6771.
Valentino Gantz, University of California, San Diego.
View Media
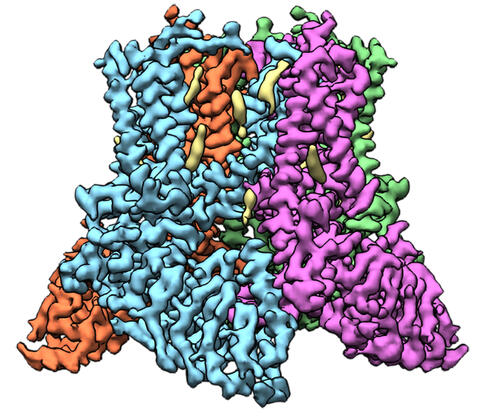
6577: Transient receptor potential channel TRPV5
6577: Transient receptor potential channel TRPV5
A 3D reconstruction of a transient receptor potential channel called TRPV5 that was created based on cryo-electron microscopy images. TRPV5 is primarily found in kidney cells and is essential for reabsorbing calcium into the blood.
Vera Moiseenkova-Bell, University of Pennsylvania.
View Media
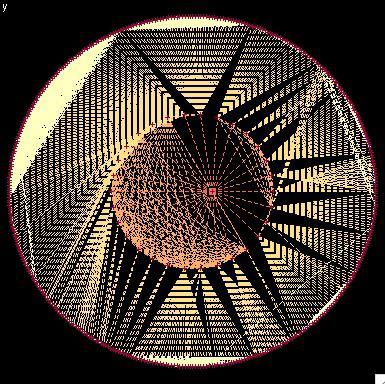
2322: Modeling disease spread
2322: Modeling disease spread
What looks like a Native American dream catcher is really a network of social interactions within a community. The red dots along the inner and outer circles represent people, while the different colored lines represent direct contact between them. All connections originate from four individuals near the center of the graph. Modeling social networks can help researchers understand how diseases spread.
Stephen Eubank, University of Virginia Biocomplexity Institute (formerly Virginia Bioinformatics Institute)
View Media
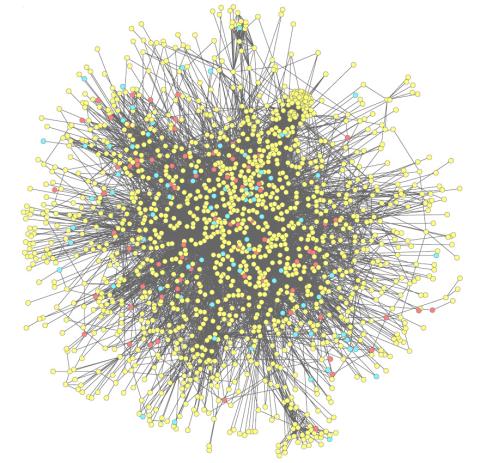
2743: Molecular interactions
2743: Molecular interactions
This network map shows molecular interactions (yellow) associated with a congenital condition that causes heart arrhythmias and the targets for drugs that alter these interactions (red and blue).
Ravi Iyengar, Mount Sinai School of Medicine
View Media
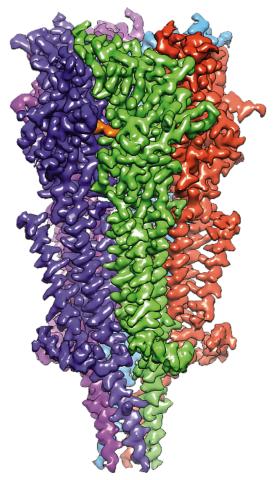
6579: Full-length serotonin receptor (ion channel)
6579: Full-length serotonin receptor (ion channel)
A 3D reconstruction, created using cryo-electron microscopy, of an ion channel known as the full-length serotonin receptor in complex with the antinausea drug granisetron (orange). Ion channels are proteins in cell membranes that help regulate many processes.
Sudha Chakrapani, Case Western Reserve University School of Medicine.
View Media
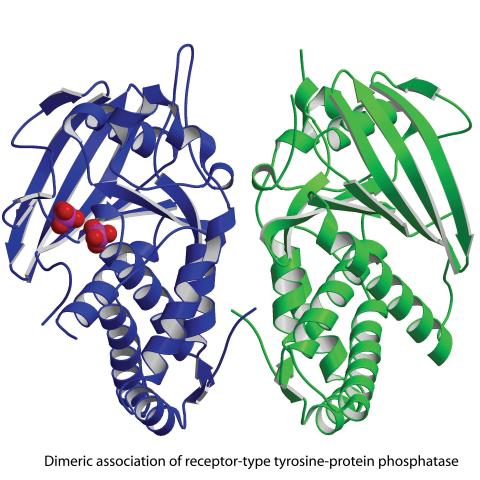
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
Model of the catalytic portion of an enzyme, receptor-type tyrosine-protein phosphatase from humans. The enzyme consists of two identical protein subunits, shown in blue and green. The groups made up of purple and red balls represent phosphate groups, chemical groups that can influence enzyme activity. This phosphatase removes phosphate groups from the enzyme tyrosine kinase, counteracting its effects.
New York Structural GenomiX Research Consortium, PSI
View Media
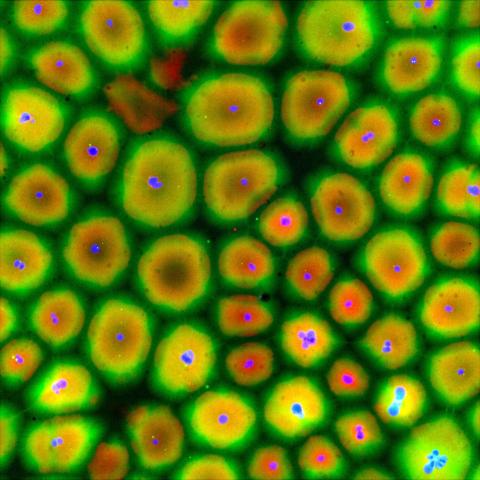
6584: Cell-like compartments from frog eggs
6584: Cell-like compartments from frog eggs
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
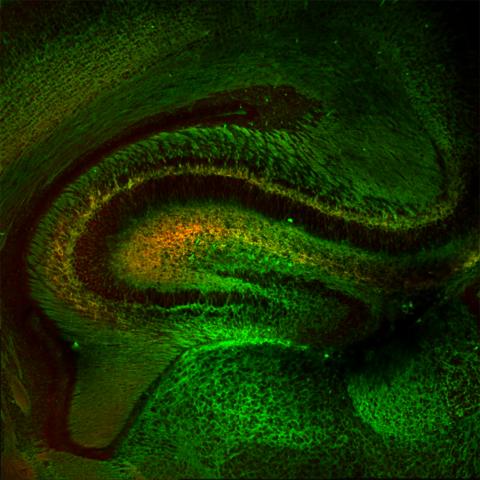
5886: Mouse Brain Cross Section
5886: Mouse Brain Cross Section
The brain sections are treated with fluorescent antibodies specific to a particular protein and visualized using serial electron microscopy (SEM).
Anton Maximov, The Scripps Research Institute, La Jolla, CA
View Media
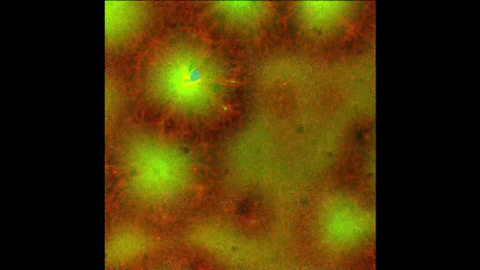
6590: Cell-like compartments emerging from scrambled frog eggs 4
6590: Cell-like compartments emerging from scrambled frog eggs 4
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Video created using confocal microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589.
Xianrui Cheng, Stanford University School of Medicine.
View Media

2543: DNA replication illustration
2543: DNA replication illustration
During DNA replication, each strand of the original molecule acts as a template for the synthesis of a new, complementary DNA strand. See image 2544 for a labeled version of this illustration.
Crabtree + Company
View Media
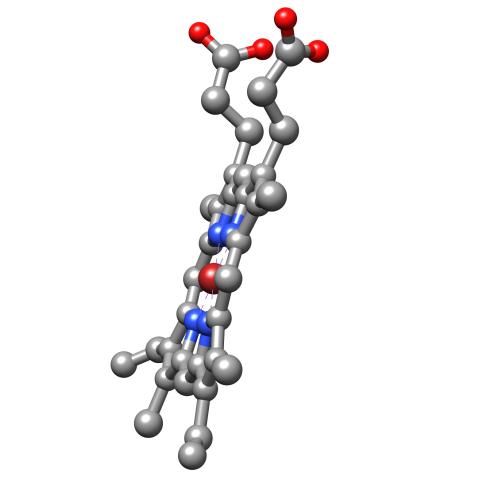
3540: Structure of heme, side view
3540: Structure of heme, side view
Molecular model of the struture of heme. Heme is a small, flat molecule with an iron ion (dark red) at its center. Heme is an essential component of hemoglobin, the protein in blood that carries oxygen throughout our bodies. This image first appeared in the September 2013 issue of Findings Magazine. View side view of heme here 3539.
Rachel Kramer Green, RCSB Protein Data Bank
View Media
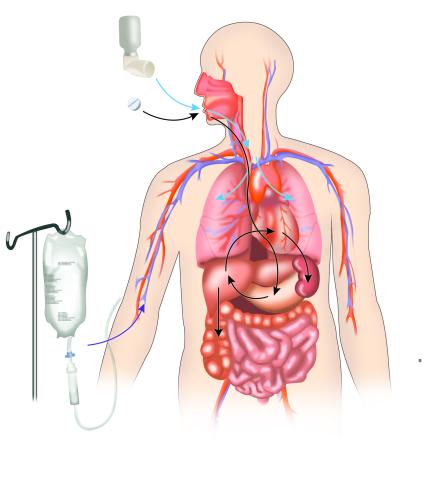
2527: A drug's life in the body
2527: A drug's life in the body
A drug's life in the body. Medicines taken by mouth pass through the liver before they are absorbed into the bloodstream. Other forms of drug administration bypass the liver, entering the blood directly. See 2528 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
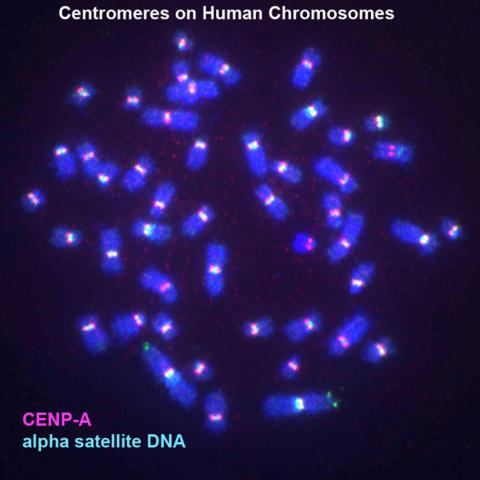
3255: Centromeres on human chromosomes
3255: Centromeres on human chromosomes
Human metaphase chromosomes are visible with fluorescence in vitro hybridization (FISH). Centromeric alpha satellite DNA (green) are found in the heterochromatin at each centromere. Immunofluorescence with CENP-A (red) shows the centromere-specific histone H3 variant that specifies the kinetochore.
Peter Warburton, Mount Sinai School of Medicine
View Media
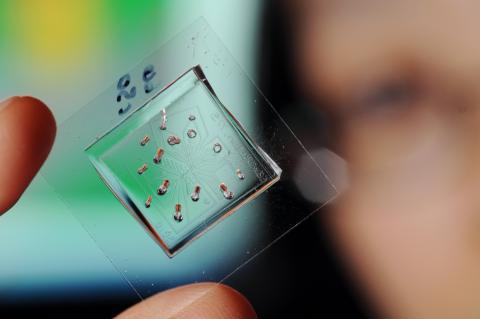
3475: Automated Worm Sorter - 4
3475: Automated Worm Sorter - 4
Georgia Tech associate professor Hang Lu holds a microfluidic chip that is part of a system that uses artificial intelligence and cutting-edge image processing to automatically examine large number of nematodes used for genetic research.
Georgia Tech/Gary Meek
View Media

2385: Heat shock protein complex from Methanococcus jannaschii
2385: Heat shock protein complex from Methanococcus jannaschii
Model based on X-ray crystallography of the structure of a small heat shock protein complex from the bacteria, Methanococcus jannaschii. Methanococcus jannaschii is an organism that lives at near boiling temperature, and this protein complex helps it cope with the stress of high temperature. Similar complexes are produced in human cells when they are "stressed" by events such as burns, heart attacks, or strokes. The complexes help cells recover from the stressful event.
Berkeley Structural Genomics Center, PSI-1
View Media
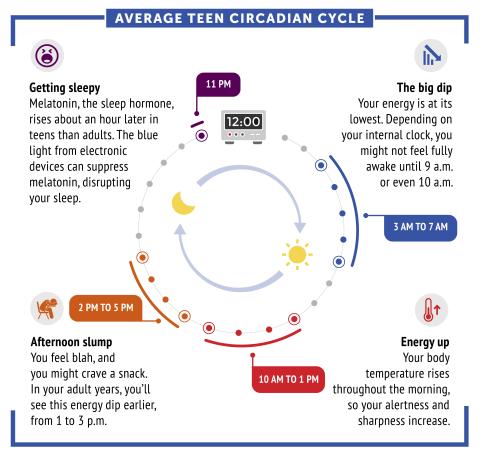
6611: Average teen circadian cycle
6611: Average teen circadian cycle
Circadian rhythms are physical, mental, and behavioral changes that follow a 24-hour cycle. Typical circadian rhythms lead to high energy during the middle of the day (10 a.m. to 1 p.m.) and an afternoon slump. At night, circadian rhythms cause the hormone melatonin to rise, making a person sleepy.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
Learn more in NIGMS’ circadian rhythms featured topics page.
See 6612 for the Spanish version of this infographic.
NIGMS
View Media
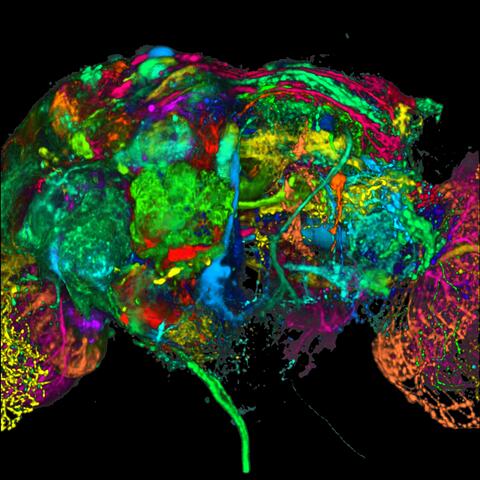
5868: Color coding of the Drosophila brain - black background
5868: Color coding of the Drosophila brain - black background
This image results from a research project to visualize which regions of the adult fruit fly (Drosophila) brain derive from each neural stem cell. First, researchers collected several thousand fruit fly larvae and fluorescently stained a random stem cell in the brain of each. The idea was to create a population of larvae in which each of the 100 or so neural stem cells was labeled at least once. When the larvae grew to adults, the researchers examined the flies’ brains using confocal microscopy. With this technique, the part of a fly’s brain that derived from a single, labeled stem cell “lights up.” The scientists photographed each brain and digitally colorized its lit-up area. By combining thousands of such photos, they created a three-dimensional, color-coded map that shows which part of the Drosophila brain comes from each of its ~100 neural stem cells. In other words, each colored region shows which neurons are the progeny or “clones” of a single stem cell. This work established a hierarchical structure as well as nomenclature for the neurons in the Drosophila brain. Further research will relate functions to structures of the brain.
Related to image 5838 and video 5843.
Related to image 5838 and video 5843.
Yong Wan from Charles Hansen’s lab, University of Utah. Data preparation and visualization by Masayoshi Ito in the lab of Kei Ito, University of Tokyo.
View Media
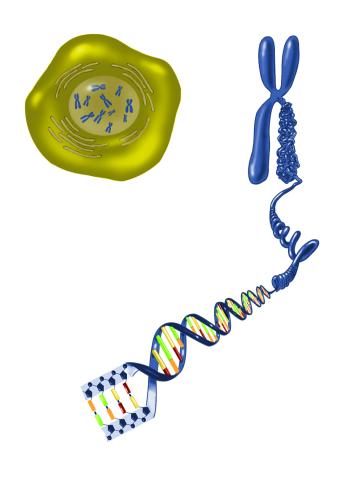
2539: Chromosome inside nucleus
2539: Chromosome inside nucleus
The long, stringy DNA that makes up genes is spooled within chromosomes inside the nucleus of a cell. (Note that a gene would actually be a much longer stretch of DNA than what is shown here.) See image 2540 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
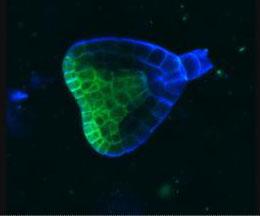
2733: Early development in Arabidopsis
2733: Early development in Arabidopsis
Early on, this Arabidopsis plant embryo picks sides: While one end will form the shoot, the other will take root underground. Short pieces of RNA in the bottom half (blue) make sure that shoot-forming genes are expressed only in the embryo's top half (green), eventually allowing a seedling to emerge with stems and leaves. Like animals, plants follow a carefully orchestrated polarization plan and errors can lead to major developmental defects, such as shoots above and below ground. Because the complex gene networks that coordinate this development in plants and animals share important similarities, studying polarity in Arabidopsis--a model organism--could also help us better understand human development.
Zachery R. Smith, Jeff Long lab at the Salk Institute for Biological Studies
View Media
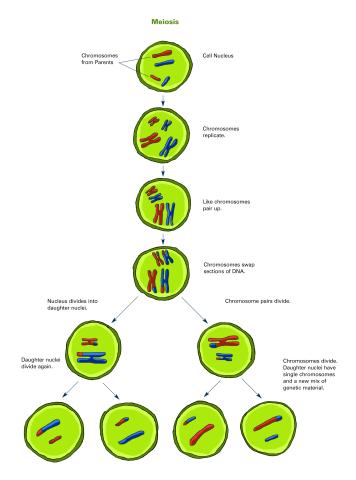
2546: Meiosis illustration (with labels)
2546: Meiosis illustration (with labels)
Meiosis is the process whereby a cell reduces its chromosomes from diploid to haploid in creating eggs or sperm. See image 2545 for an unlabeled version of this illustration. See image 2544 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
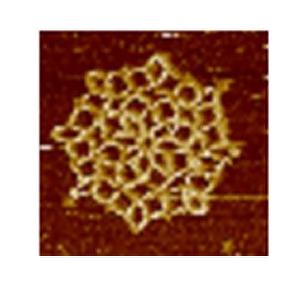
3724: Snowflake DNA origami
3724: Snowflake DNA origami
An atomic force microscopy image shows DNA folded into an intricate, computer-designed structure. The image is featured on Biomedical Beat blog post Cool Images: A Holiday-Themed Collection. For more background on DNA origami, see Cool Image: DNA Origami. See also related image 3690.
Hao Yan, Arizona State University
View Media
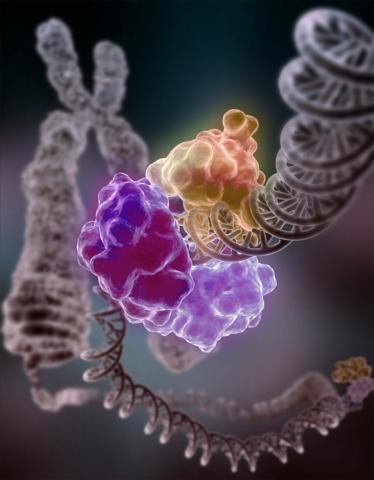
3493: Repairing DNA
3493: Repairing DNA
Like a watch wrapped around a wrist, a special enzyme encircles the double helix to repair a broken strand of DNA. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. Related to image 2330.
Tom Ellenberger, Washington University School of Medicine
View Media
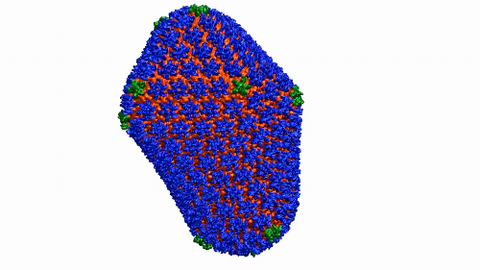
6601: Atomic-level structure of the HIV capsid
6601: Atomic-level structure of the HIV capsid
This animation shows atoms of the HIV capsid, the shell that encloses the virus's genetic material. Scientists determined the exact structure of the capsid using a variety of imaging techniques and analyses. They then entered this data into a supercomputer to produce this image. Related to image 3477.
Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
View Media
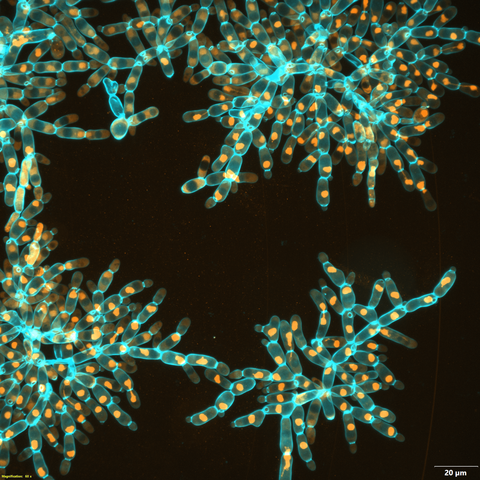
6971: Snowflake yeast 3
6971: Snowflake yeast 3
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Here, the researchers visualized nuclei in orange to help them study changes in how the yeast cells divided. Cell walls are shown in blue. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6970.
Related to images 6969 and 6970.
William Ratcliff, Georgia Institute of Technology.
View Media
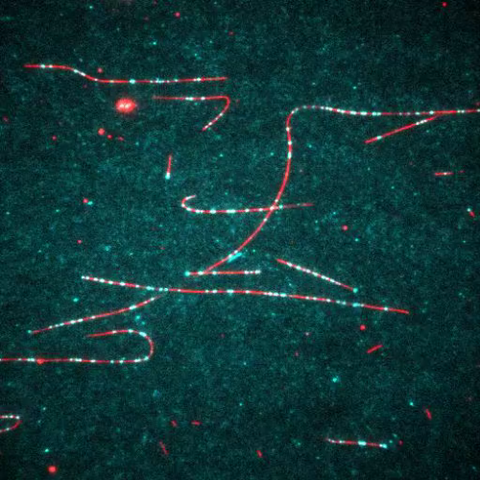
7023: Dynein moving along microtubules
7023: Dynein moving along microtubules
Dynein (green) is a motor protein that “walks” along microtubules (red, part of the cytoskeleton) and carries its cargo along with it. This video was captured through fluorescence microscopy.
Morgan DeSantis, University of Michigan.
View Media
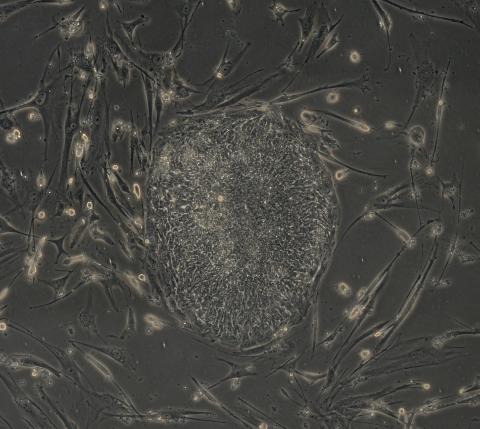
2606: Induced stem cells from adult skin 04
2606: Induced stem cells from adult skin 04
The human skin cells pictured contain genetic modifications that make them pluripotent, essentially equivalent to embryonic stem cells. A scientific team from the University of Wisconsin-Madison including researchers Junying Yu, James Thomson, and their colleagues produced the transformation by introducing a set of four genes into human fibroblasts, skin cells that are easy to obtain and grow in culture.
James Thomson, University of Wisconsin-Madison
View Media
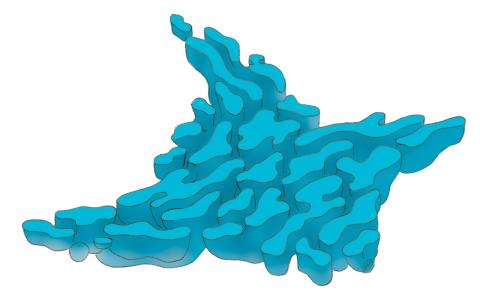
1292: Smooth ER
1292: Smooth ER
The endoplasmic reticulum comes in two types: Rough ER is covered with ribosomes and prepares newly made proteins; smooth ER specializes in making lipids and breaking down toxic molecules.
Judith Stoffer
View Media
3786: Movie of in vitro assembly of a cell-signaling pathway
3786: Movie of in vitro assembly of a cell-signaling pathway
T cells are white blood cells that are important in defending the body against bacteria, viruses and other pathogens. Each T cell carries proteins, called T-cell receptors, on its surface that are activated when they come in contact with an invader. This activation sets in motion a cascade of biochemical changes inside the T cell to mount a defense against the invasion. Scientists have been interested for some time what happens after a T-cell receptor is activated. One obstacle has been to study how this signaling cascade, or pathway, proceeds inside T cells.
In this video, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The video shows three key steps during the signaling process: phosphorylation of the T-cell receptor (green), clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to image 3787.
In this video, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The video shows three key steps during the signaling process: phosphorylation of the T-cell receptor (green), clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to image 3787.
Xiaolei Su, HHMI Whitman Center of the Marine Biological Laboratory
View Media
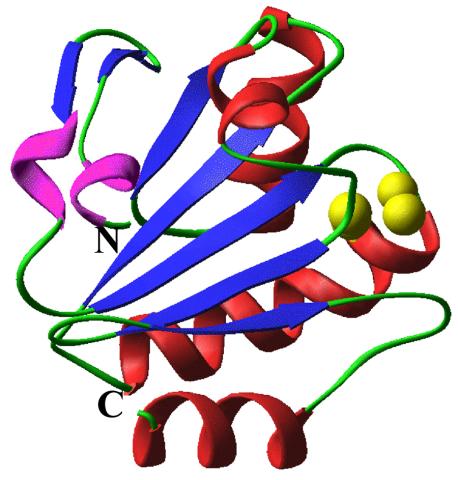
2382: PanB from M. tuberculosis (2)
2382: PanB from M. tuberculosis (2)
Model of an enzyme, PanB, from Mycobacterium tuberculosis, the bacterium that causes most cases of tuberculosis. This enzyme is an attractive drug target.
Mycobacterium Tuberculosis Center, PSI-1
View Media
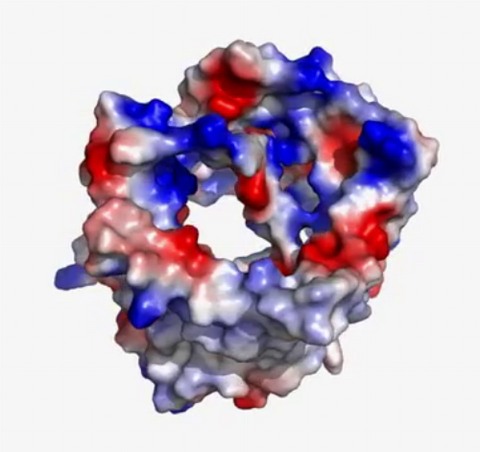
2571: VDAC video 02
2571: VDAC video 02
This video shows the structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to videos 2570 and 2572.
Related to videos 2570 and 2572.
Gerhard Wagner, Harvard Medical School
View Media

3719: CRISPR illustration
3719: CRISPR illustration
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and download the four images of the CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and download the four images of the CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
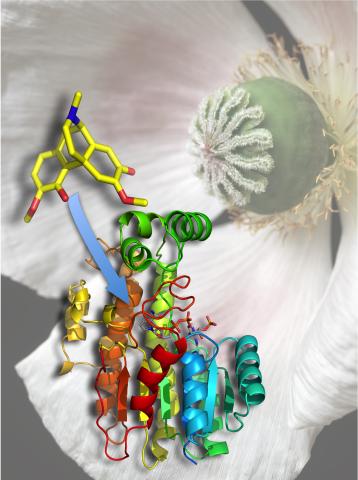
3422: Atomic Structure of Poppy Enzyme
3422: Atomic Structure of Poppy Enzyme
The atomic structure of the morphine biosynthetic enzyme salutaridine reductase bound to the cofactor NADPH. The substrate salutaridine is shown entering the active site.
Judy Coyle, Donald Danforth Plant Science Center
View Media
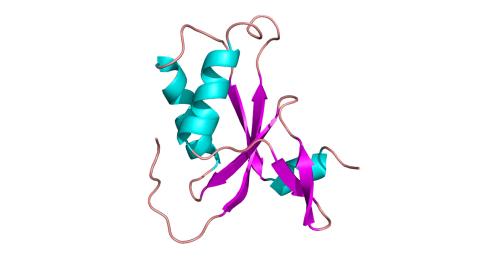
3427: Antitoxin GhoS (Illustration 1)
3427: Antitoxin GhoS (Illustration 1)
Structure of the bacterial antitoxin protein GhoS. GhoS inhibits the production of a bacterial toxin, GhoT, which can contribute to antibiotic resistance. GhoS is the first known bacterial antitoxin that works by cleaving the messenger RNA that carries the instructions for making the toxin. More information can be found in the paper: Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012 Oct;8(10):855-61. Related to 3428.
Rebecca Page and Wolfgang Peti, Brown University and Thomas K. Wood, Pennsylvania State University
View Media
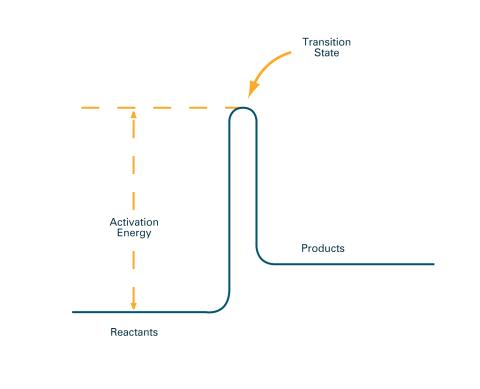
2526: Activation energy (with labels)
2526: Activation energy (with labels)
To become products, reactants must overcome an energy hill. See image 2525 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
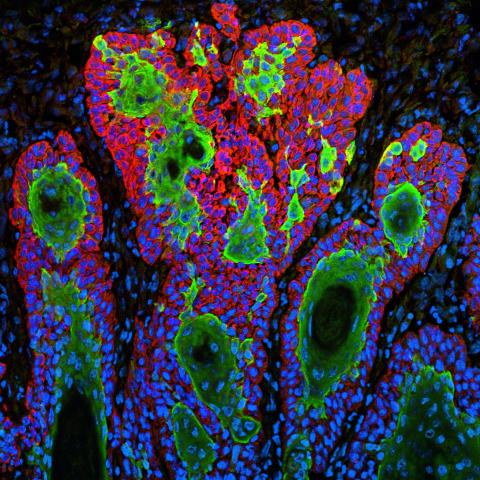
3628: Skin cancer cells (squamous cell carcinoma)
3628: Skin cancer cells (squamous cell carcinoma)
This image shows the uncontrolled growth of cells in squamous cell carcinoma, the second most common form of skin cancer. If caught early, squamous cell carcinoma is usually not life-threatening.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Markus Schober and Elaine Fuchs, The Rockefeller University
View Media
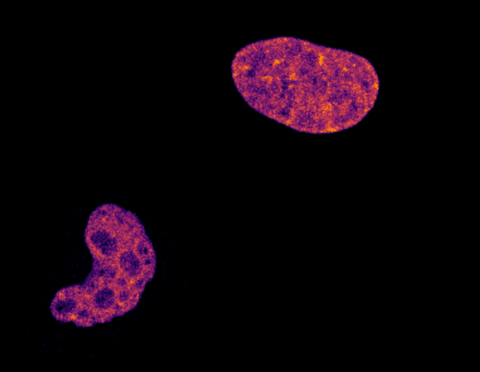
6790: Cell division and cell death
6790: Cell division and cell death
Two cells over a 2-hour period. The one on the bottom left goes through programmed cell death, also known as apoptosis. The one on the top right goes through cell division, also called mitosis. This video was captured using a confocal microscope.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media