Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.

2779: Mature, flowering Arabidopsis
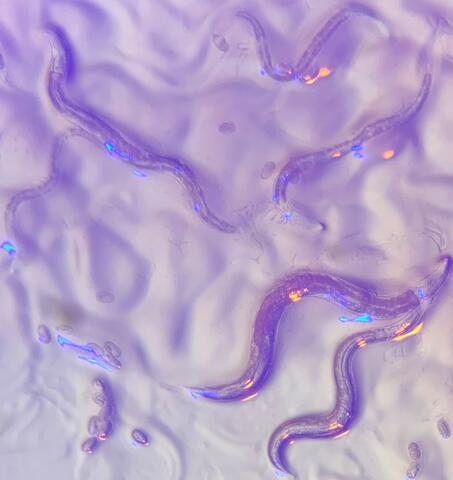
6750: C. elegans with blue and yellow lights in the background
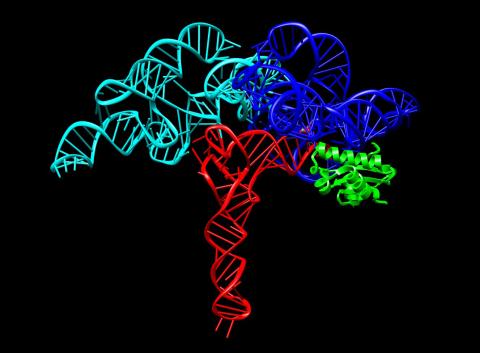
3660: Ribonuclease P structure

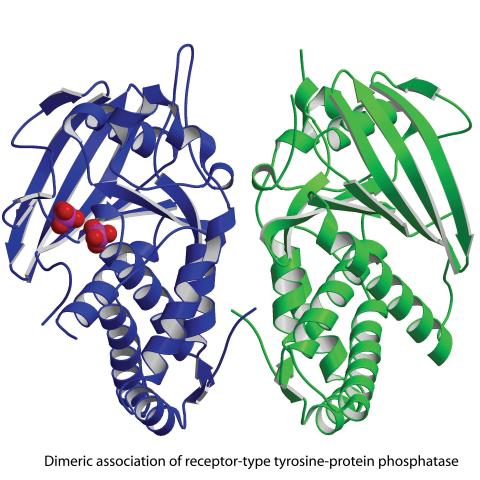
2349: Dimeric association of receptor-type tyrosine-protein phosphatase

1294: Stem cell differentiation

2395: Fungal lipase (1)
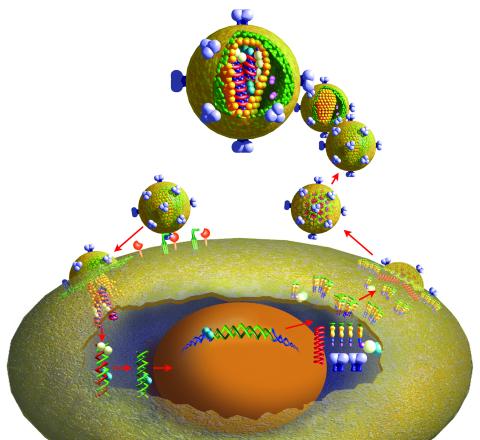
2513: Life of an AIDS virus
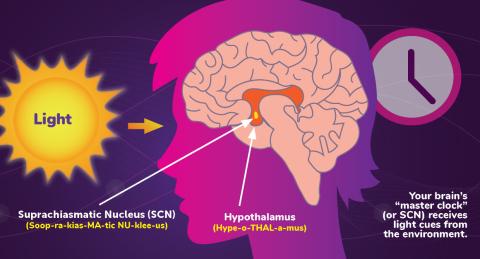
6613: Circadian rhythms and the SCN
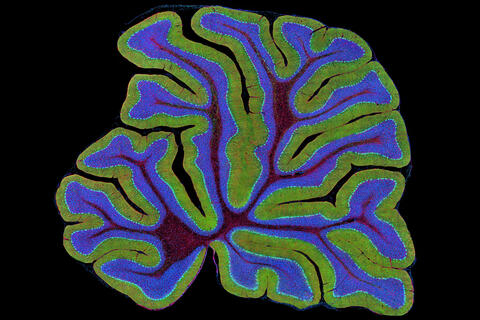
3639: Cerebellum: the brain's locomotion control center
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.

7011: Hawaiian bobtail squid
Related to image 7010 and video 7012.
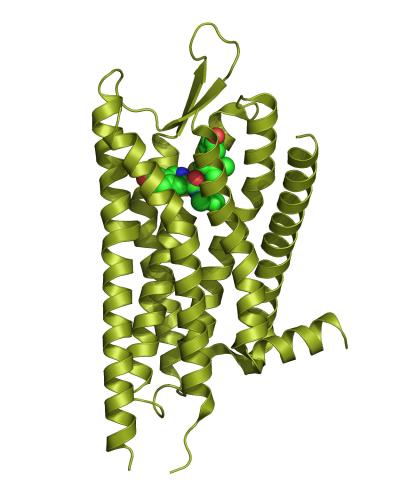
3359: Kappa opioid receptor
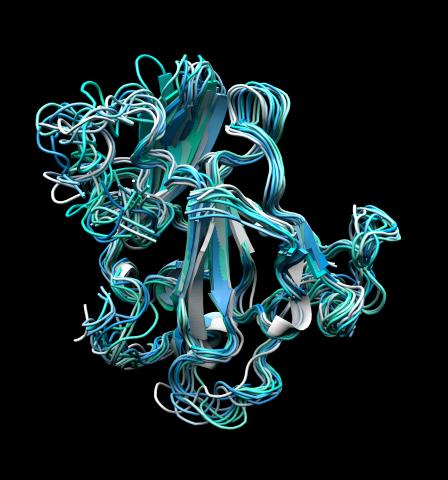
5866: Structure of a key antigen protein involved with Hepatitis C Virus infection
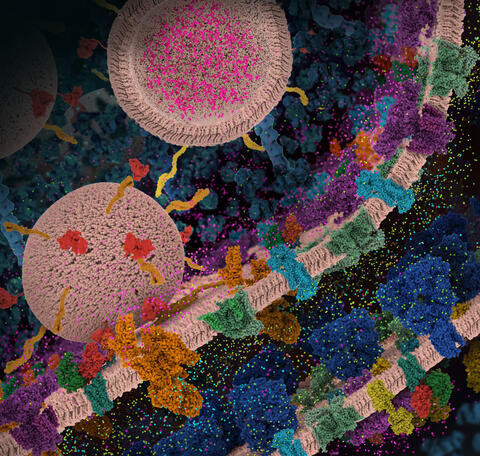
6992: Molecular view of glutamatergic synapse
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
6562: Drosophila (fruit fly) myosin 1D motility assay
2796: Anti-tumor drug ecteinascidin 743 (ET-743), structure without hydrogens 03
6589: Cell-like compartments emerging from scrambled frog eggs 3
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, and 6590.
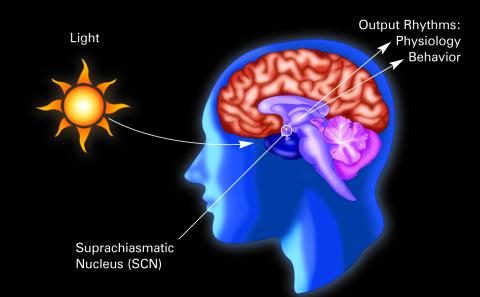
2569: Circadian rhythm (with labels)
6520: HeLa cell undergoing division into two daughter cells
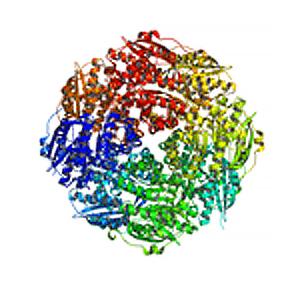
2350: Mandelate racemase from B. subtilis
5760: Annotated TEM cross-section of C. elegans (roundworm)
The image is from a figure in an article published in the journal eLife.

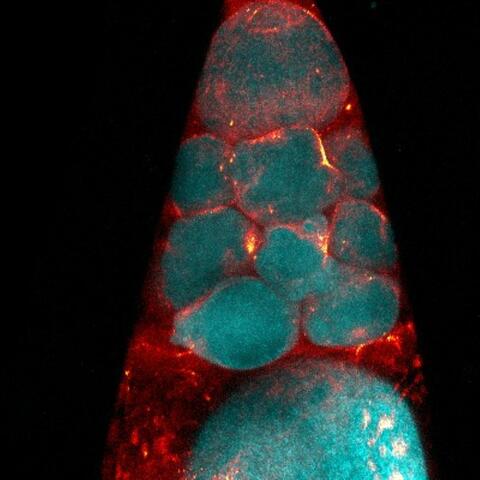
3723: Fluorescent microscopy of kidney tissue
Related to entries 3725 and 3675.
6556: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 72 hour
See 6557 for a photo of this process at 24 hours on 0.75% agar surface.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6550 for a video of this process.
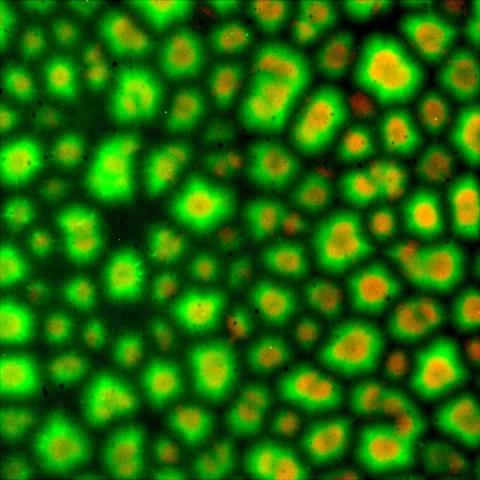
6586: Cell-like compartments from frog eggs 3
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
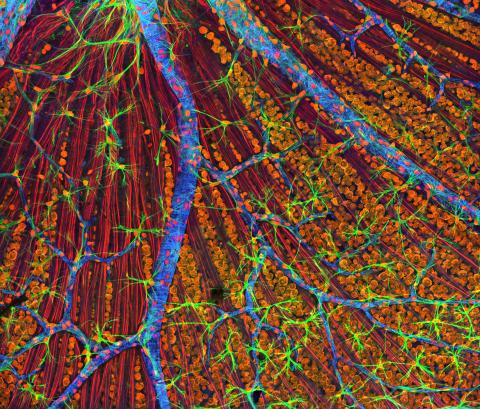
3309: Mouse Retina

2443: Mapping human genetic variation
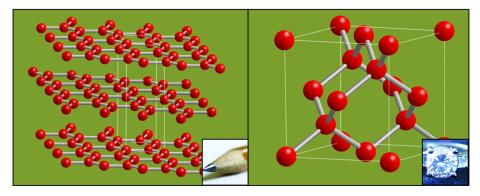
2507: Carbon building blocks (with examples)

1283: Vesicle traffic

6549: The Structure of Cilia’s Doublet Microtubules
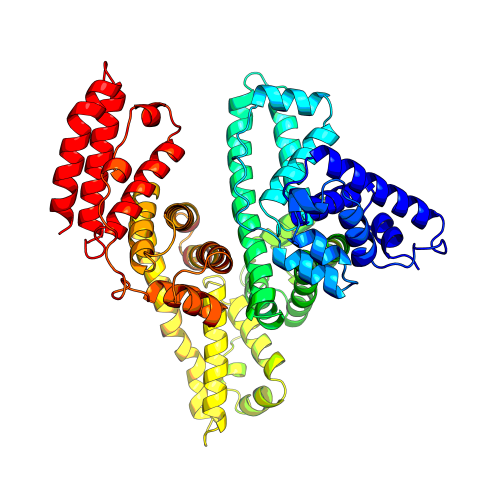
3745: Serum albumin structure 2
Related to entries 3744 and 3746
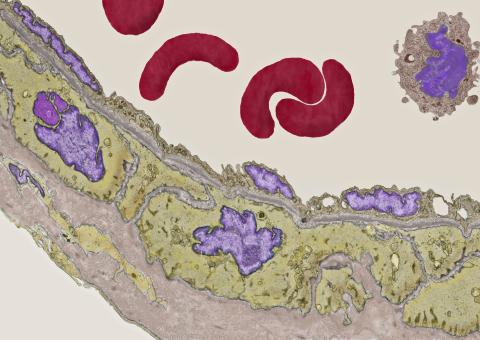
3738: Transmission electron microscopy of coronary artery wall with elastin-rich ECM pseudocolored in light brown
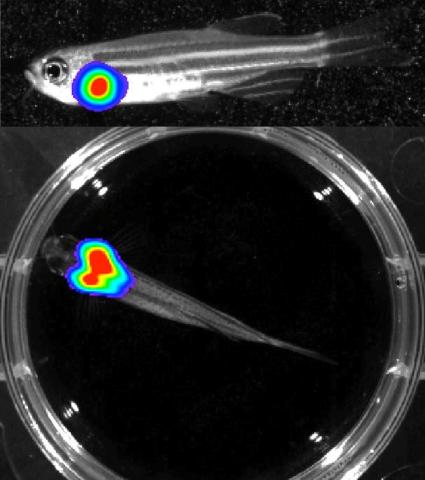
3556: Bioluminescent imaging in adult zebrafish - lateral and overhead view
For imagery of the overhead view go to 3557.
For imagery of the lateral view go to 3558.
For more information about the illumated area go to 3559.
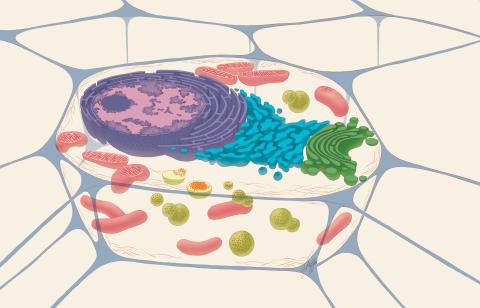
1274: Animal cell
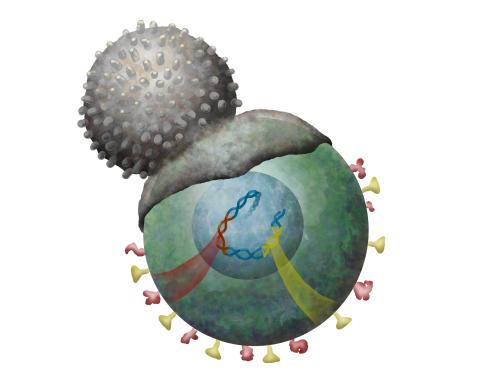
2489: Immune cell attacks cell infected with a retrovirus
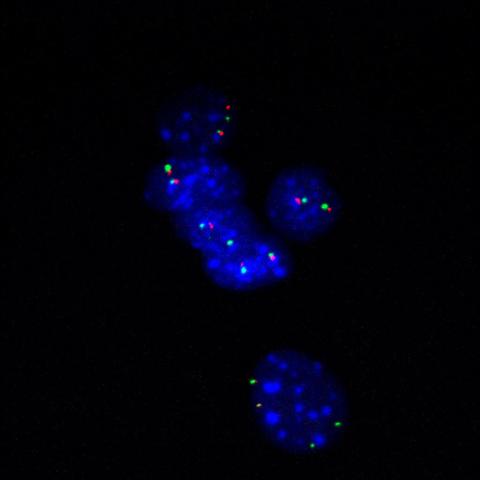
3296: Fluorescence in situ hybridization (FISH) in mouse ES cells shows DNA interactions
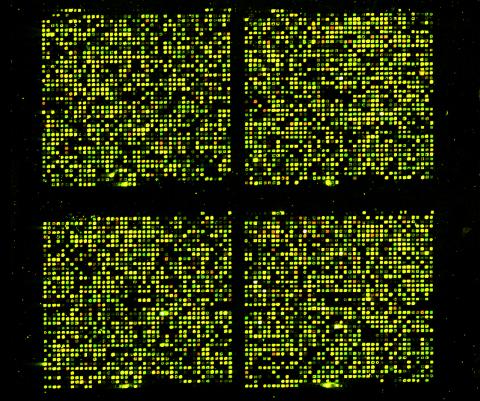
1070: Microarray 01

1335: Telomerase illustration
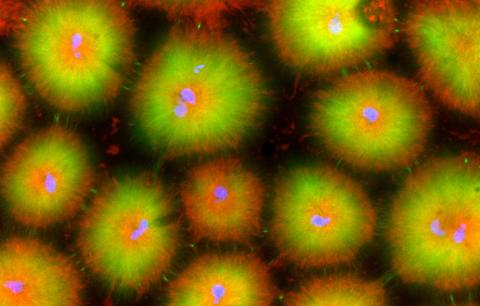
6591: Cell-like compartments from frog eggs 4
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
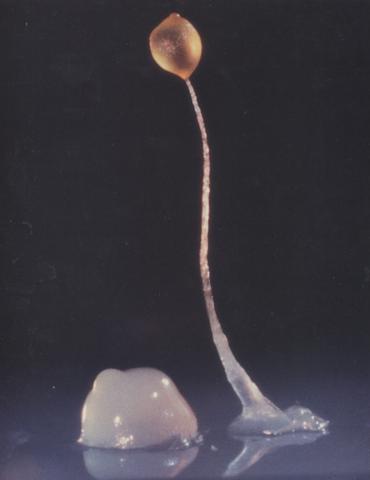
2684: Dicty fruit

1060: Protein crystals
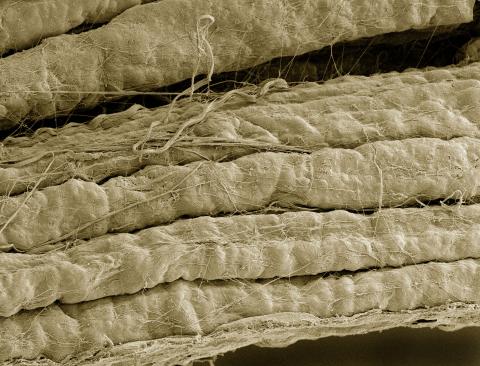
3737: A bundle of myelinated peripheral nerve cells (axons)
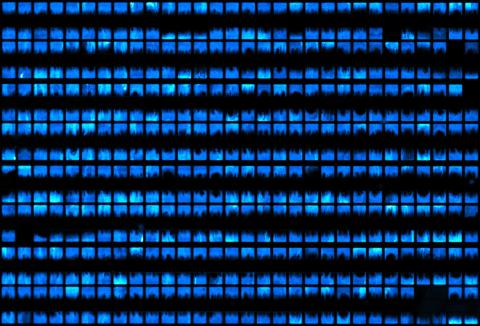
3266: Biopixels
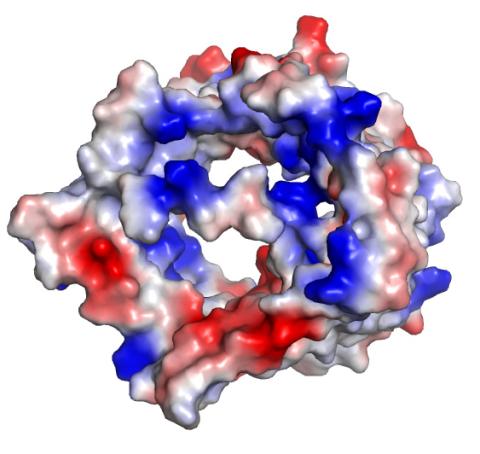
2491: VDAC-1 (2)
Related to images 2494, 2495, and 2488.
1087: Natcher Building 07
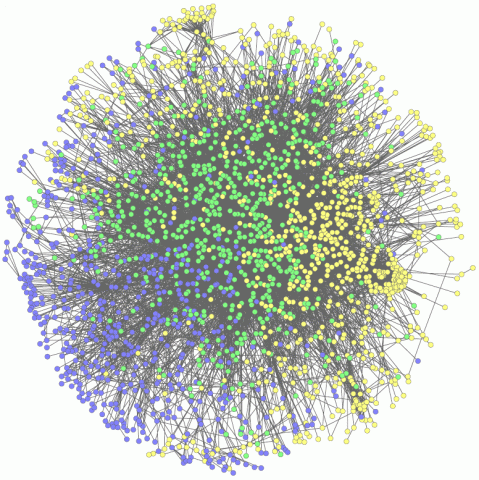
2735: Network Map

3530: Lorsch Swearing In
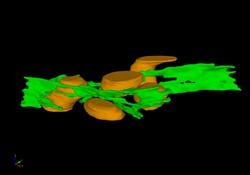
2635: Mitochondria and endoplasmic reticulum
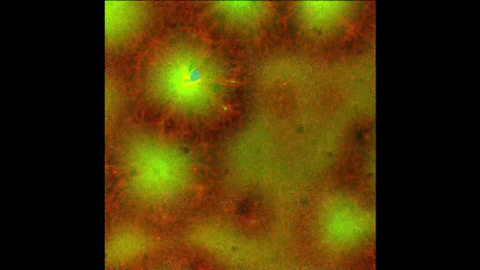
6590: Cell-like compartments emerging from scrambled frog eggs 4
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589.
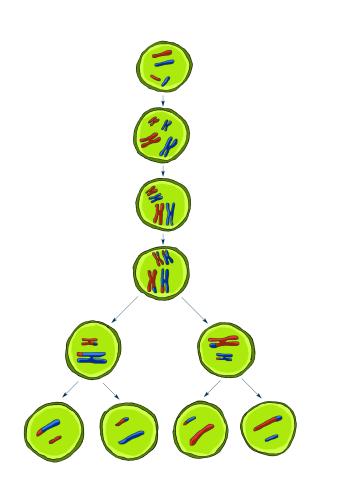
2545: Meiosis illustration