Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
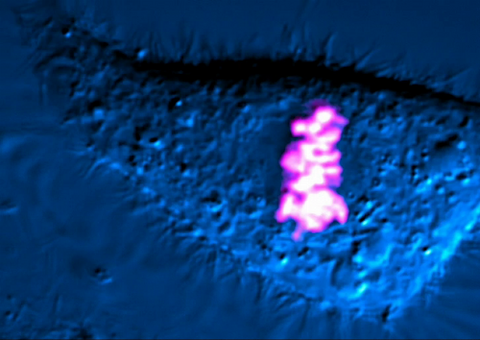
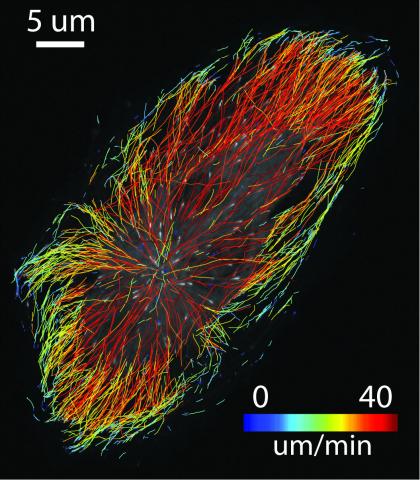
6965: Dividing cell
6965: Dividing cell
As this cell was undergoing cell division, it was imaged with two microscopy techniques: differential interference contrast (DIC) and confocal. The DIC view appears in blue and shows the entire cell. The confocal view appears in pink and shows the chromosomes.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media
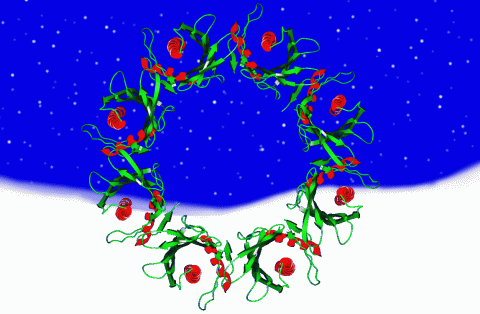
2372: Wreath-shaped protein from X. campestris
2372: Wreath-shaped protein from X. campestris
Crystal structure of a protein with unknown function from Xanthomonas campestris, a plant pathogen. Eight copies of the protein crystallized to form a ring. Chosen as the December 2007 Protein Structure Initiative Structure of the Month.
Ken Schwinn and Sonia Espejon-Reynes, New York SGX Research Center for Structural Genomics
View Media
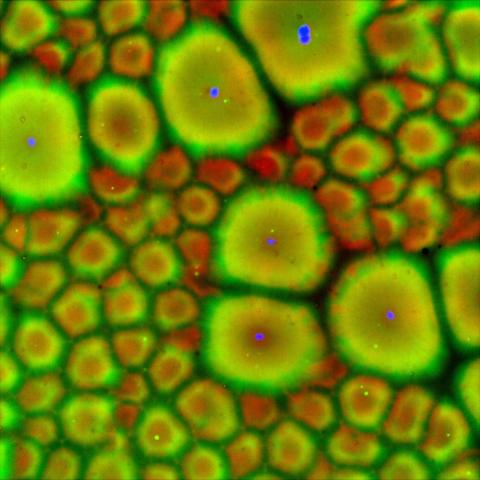
6585: Cell-like compartments from frog eggs 2
6585: Cell-like compartments from frog eggs 2
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Regions without nuclei formed smaller compartments. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
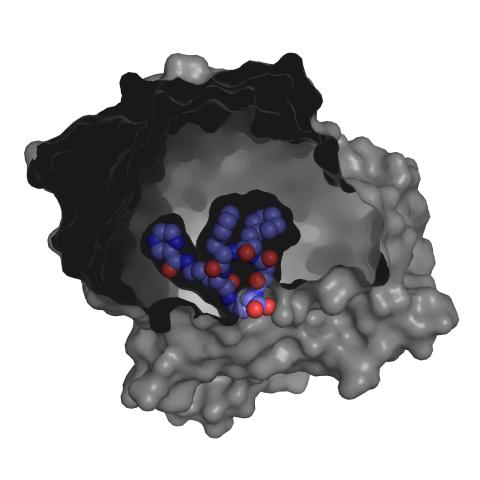
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
3415: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 3
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to 3413, 3414, 3416, 3417, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
1087: Natcher Building 07
1087: Natcher Building 07
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
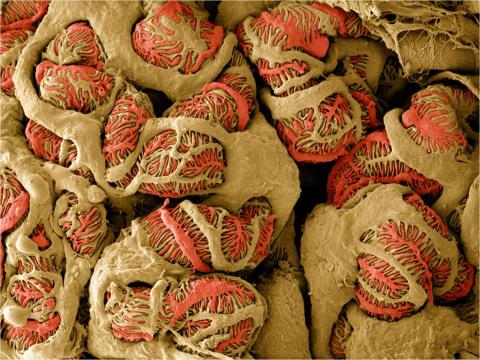
3392: NCMIR Kidney Glomeruli
3392: NCMIR Kidney Glomeruli
Stained glomeruli in the kidney. The kidney is an essential organ responsible for disposing wastes from the body and for maintaining healthy ion levels in the blood. It works like a purifier by pulling break-down products of metabolism, such as urea and ammonium, from the bloodstream for excretion in urine. The glomerulus is a structure that helps filter the waste compounds from the blood. It consists of a network of capillaries enclosed within a Bowman's capsule of a nephron, which is the structure in which ions exit or re-enter the blood in the kidney.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
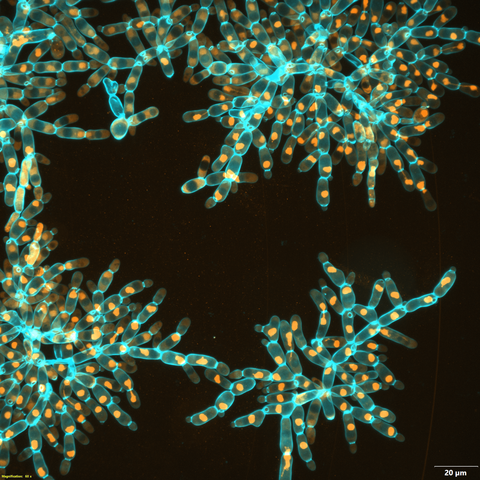
6971: Snowflake yeast 3
6971: Snowflake yeast 3
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Here, the researchers visualized nuclei in orange to help them study changes in how the yeast cells divided. Cell walls are shown in blue. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6970.
Related to images 6969 and 6970.
William Ratcliff, Georgia Institute of Technology.
View Media
6540: Pathways: What is It? | Why Scientists Study Cells
6540: Pathways: What is It? | Why Scientists Study Cells
Learn how curiosity about the world and our cells is key to scientific discoveries. Discover more resources from NIGMS’ Pathways collaboration with Scholastic. View the video on YouTube for closed captioning.
National Institute of General Medical Sciences
View Media
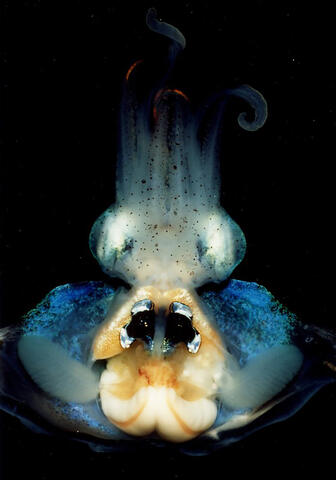
7013: An adult Hawaiian bobtail squid
7013: An adult Hawaiian bobtail squid
An adult female Hawaiian bobtail squid, Euprymna scolopes, with its mantle cavity exposed from the underside. Some internal organs are visible, including the two lobes of the light organ that contains bioluminescent bacteria, Vibrio fischeri. The light organ includes accessory tissues like an ink sac (black) that serves as a shutter, and a silvery reflector that directs the light out of the underside of the animal.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
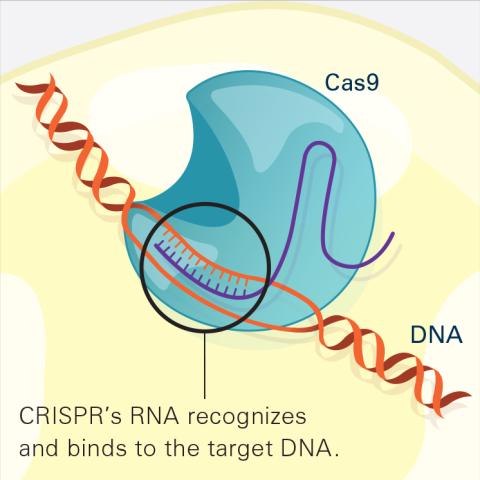
6486: CRISPR Illustration Frame 2
6486: CRISPR Illustration Frame 2
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). In this frame (2 of 4), the CRISPR machine locates the target DNA sequence once inserted into a cell.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
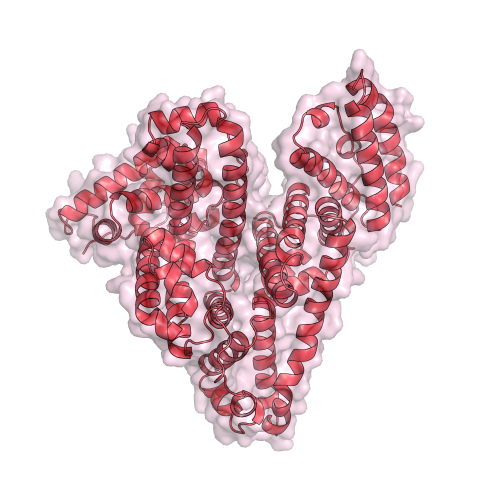
3744: Serum albumin structure 1
3744: Serum albumin structure 1
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3745 and 3746.
Related to entries 3745 and 3746.
Wladek Minor, University of Virginia
View Media
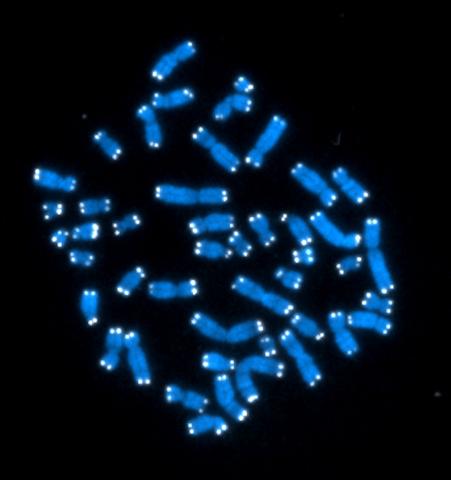
2626: Telomeres
2626: Telomeres
The 46 human chromosomes are shown in blue, with the telomeres appearing as white pinpoints. The DNA has already been copied, so each chromosome is actually made up of two identical lengths of DNA, each with its own two telomeres.
Hesed Padilla-Nash and Thomas Ried, the National Cancer Institute, a part of NIH
View Media
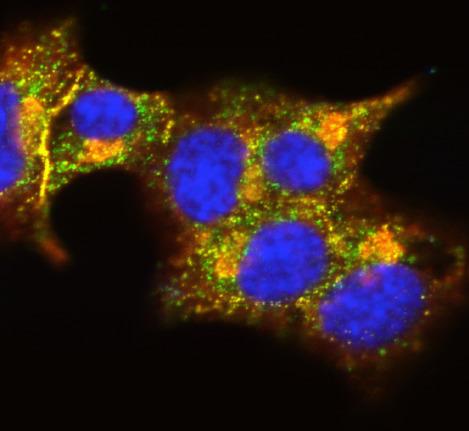
3546: Insulin and protein interact in pancreatic beta cells
3546: Insulin and protein interact in pancreatic beta cells
A large number of proteins interact with the hormone insulin as it is produced in and secreted from the beta cells of the pancreas. In this image, the interactions of TMEM24 protein (green) and insulin (red) in pancreatic beta cells are shown in yellow. More information about the research behind this image can be found in a Biomedical Beat Blog posting from November 2013.
William E. Balch, The Scripps Research Institute
View Media
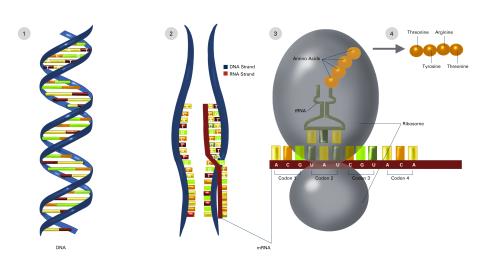
2549: Central dogma, illustrated (with labels and numbers for stages)
2549: Central dogma, illustrated (with labels and numbers for stages)
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a version of this illustration that isn't numbered and 2547 for a an entirely unlabeled version. Featured in The New Genetics.
Crabtree + Company
View Media
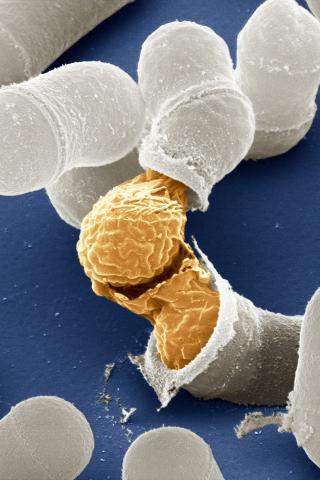
3614: Birth of a yeast cell
3614: Birth of a yeast cell
Yeast make bread, beer, and wine. And like us, yeast can reproduce sexually. A mother and father cell fuse and create one large cell that contains four offspring. When environmental conditions are favorable, the offspring are released, as shown here. Yeast are also a popular study subject for scientists. Research on yeast has yielded vast knowledge about basic cellular and molecular biology as well as about myriad human diseases, including colon cancer and various metabolic disorders.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Juergen Berger, Max Planck Institute for Developmental Biology, and Maria Langegger, Friedrich Miescher Laboratory of the Max Planck Society, Germany
View Media
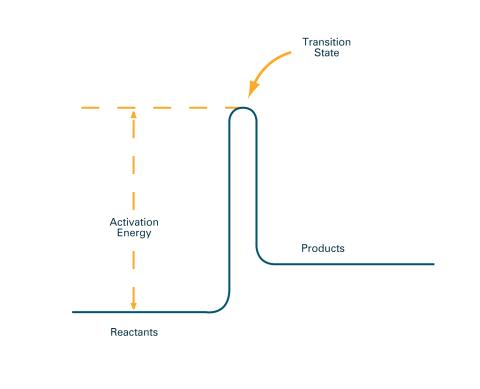
2526: Activation energy (with labels)
2526: Activation energy (with labels)
To become products, reactants must overcome an energy hill. See image 2525 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media

2375: Protein purification robot
2375: Protein purification robot
Irina Dementieva, a biochemist, and Youngchang Kim, a biophysicist and crystallographer, work with the first robot of its type in the U.S. to automate protein purification. The robot, which is housed in a refrigerator, is an integral part of the Midwest Structural Genomics Center's plan to automate the protein crystallography process.
Midwest Center for Structural Genomics
View Media
3788: Yeast cells pack a punch
3788: Yeast cells pack a punch
Although they are tiny, microbes that are growing in confined spaces can generate a lot of pressure. In this video, yeast cells grow in a small chamber called a microfluidic bioreactor. As the cells multiply, they begin to bump into and squeeze each other, resulting in periodic bursts of cells moving into different parts of the chamber. The continually growing cells also generate a lot of pressure--the researchers conducting these experiments found that the pressure generated by the cells can be almost five times higher than that in a car tire--about 150 psi, or 10 times the atmospheric pressure. Occasionally, this pressure even caused the small reactor to burst. By tracking the growth of the yeast or other cells and measuring the mechanical forces generated, scientists can simulate microbial growth in various places such as water pumps, sewage lines or catheters to learn how damage to these devices can be prevented. To learn more how researchers used small bioreactors to gauge the pressure generated by growing microbes, see this press release from UC Berkeley.
Oskar Hallatschek, UC Berkeley
View Media

2667: Glowing fish
2667: Glowing fish
Professor Marc Zimmer's family pets, including these fish, glow in the dark in response to blue light. Featured in the September 2009 issue of Findings.
View Media

3730: A molecular interaction network in yeast 1
3730: A molecular interaction network in yeast 1
The image visualizes a part of the yeast molecular interaction network. The lines in the network represent connections among genes (shown as little dots) and different-colored networks indicate subnetworks, for instance, those in specific locations or pathways in the cell. Researchers use gene or protein expression data to build these networks; the network shown here was visualized with a program called Cytoscape. By following changes in the architectures of these networks in response to altered environmental conditions, scientists can home in on those genes that become central "hubs" (highly connected genes), for example, when a cell encounters stress. They can then further investigate the precise role of these genes to uncover how a cell's molecular machinery deals with stress or other factors. Related to images 3732 and 3733.
Keiichiro Ono, UCSD
View Media

3755: Cryo-EM reveals how the HIV capsid attaches to a human protein to evade immune detection
3755: Cryo-EM reveals how the HIV capsid attaches to a human protein to evade immune detection
The illustration shows the capsid of human immunodeficiency virus (HIV) whose molecular features were resolved with cryo-electron microscopy (cryo-EM). On the left, the HIV capsid is "naked," a state in which it would be easily detected by and removed from cells. However, as shown on the right, when the viral capsid binds to and is covered with a host protein, called cyclophilin A (shown in red), it evades detection and enters and invades the human cell to use it to establish an infection. To learn more about how cyclophilin A helps HIV infect cells and how scientists used cryo-EM to find out the mechanism by which the HIV capsid attaches to cyclophilin A, see this news release by the University of Illinois. A study reporting these findings was published in the journal Nature Communications.
Juan R. Perilla, University of Illinois at Urbana-Champaign
View Media
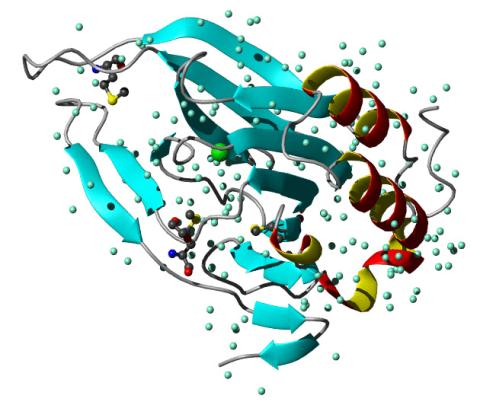
2347: Cysteine dioxygenase from mouse
2347: Cysteine dioxygenase from mouse
Model of the mammalian iron enzyme cysteine dioxygenase from a mouse.
Center for Eukaryotic Structural Genomics, PSI
View Media
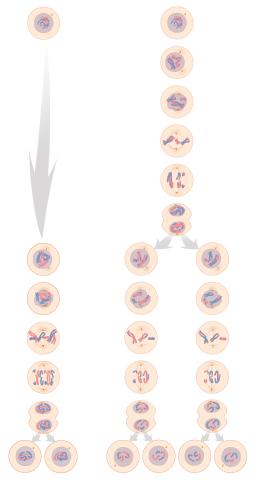
1333: Mitosis and meiosis compared
1333: Mitosis and meiosis compared
Meiosis is used to make sperm and egg cells. During meiosis, a cell's chromosomes are copied once, but the cell divides twice. During mitosis, the chromosomes are copied once, and the cell divides once. For simplicity, cells are illustrated with only three pairs of chromosomes. See image 6788 for a labeled version of this illustration.
Judith Stoffer
View Media
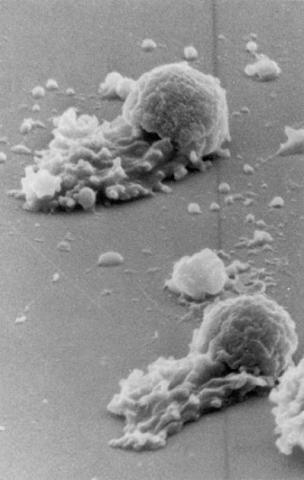
3489: Worm sperm
3489: Worm sperm
To develop a system for studying cell motility in unnatrual conditions -- a microscope slide instead of the body -- Tom Roberts and Katsuya Shimabukuro at Florida State University disassembled and reconstituted the motility parts used by worm sperm cells.
Tom Roberts, Florida State University
View Media
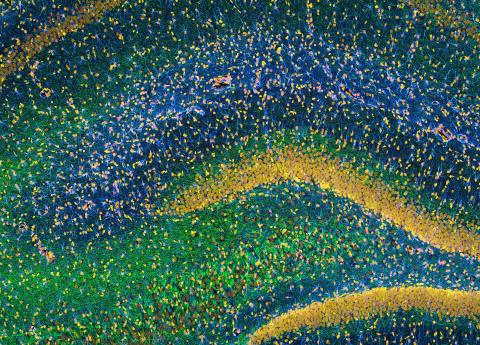
3308: Rat Hippocampus
3308: Rat Hippocampus
This image of the hippocampus was taken with an ultra-widefield high-speed multiphoton laser microscope. Tissue was stained to reveal the organization of glial cells (cyan), neurofilaments (green) and DNA (yellow). The microscope Deerinck used was developed in conjunction with Roger Tsien (2008 Nobel laureate in Chemistry) and remains a powerful and unique tool today.
Tom Deerinck, NCMIR
View Media
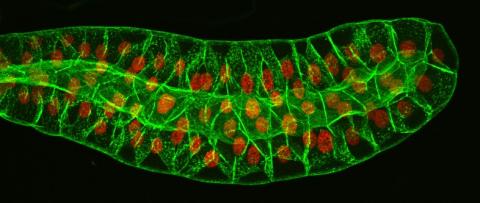
3603: Salivary gland in the developing fruit fly
3603: Salivary gland in the developing fruit fly
For fruit flies, the salivary gland is used to secrete materials for making the pupal case, the protective enclosure in which a larva transforms into an adult fly. For scientists, this gland provided one of the earliest glimpses into the genetic differences between individuals within a species. Chromosomes in the cells of these salivary glands replicate thousands of times without dividing, becoming so huge that scientists can easily view them under a microscope and see differences in genetic content between individuals.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Richard Fehon, University of Chicago
View Media
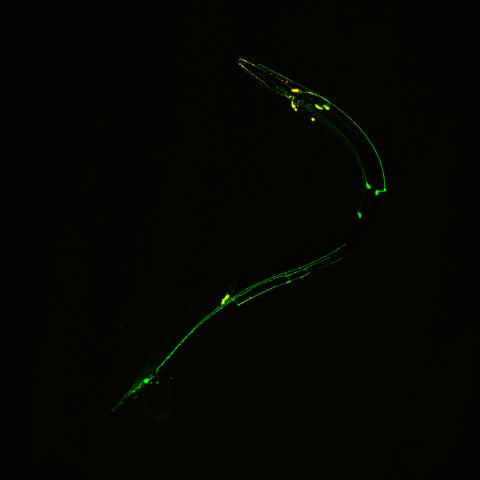
3252: Neural circuits in worms similar to those in humans
3252: Neural circuits in worms similar to those in humans
Green and yellow fluorescence mark the processes and cell bodies of some C. elegans neurons. Researchers have found that the strategies used by this tiny roundworm to control its motions are remarkably similar to those used by the human brain to command movement of our body parts. From a November 2011 University of Michigan news release.
Shawn Xu, University of Michigan
View Media
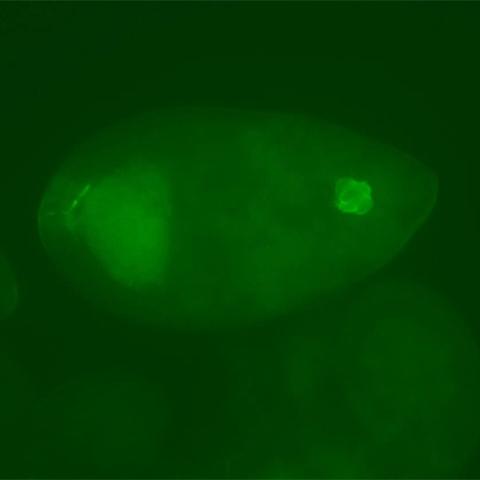
3594: Fly cells
3594: Fly cells
If a picture is worth a thousand words, what's a movie worth? For researchers studying cell migration, a "documentary" of fruit fly cells (bright green) traversing an egg chamber could answer longstanding questions about cell movement. See 2315 for video.
Denise Montell, Johns Hopkins University School of Medicine
View Media

2319: Mapping metabolic activity
2319: Mapping metabolic activity
Like a map showing heavily traveled roads, this mathematical model of metabolic activity inside an E. coli cell shows the busiest pathway in white. Reaction pathways used less frequently by the cell are marked in red (moderate activity) and green (even less activity). Visualizations like this one may help scientists identify drug targets that block key metabolic pathways in bacteria.
Albert-László Barabási, University of Notre Dame
View Media
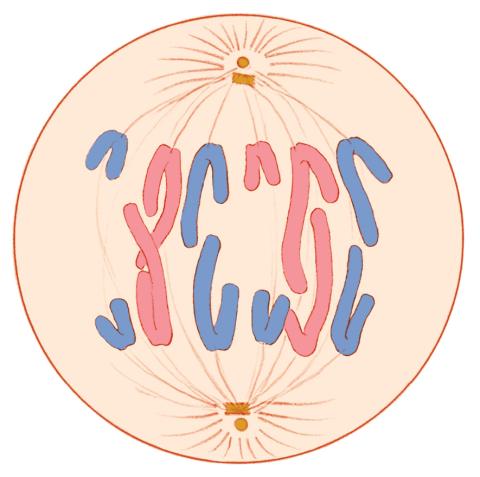
1328: Mitosis - anaphase
1328: Mitosis - anaphase
A cell in anaphase during mitosis: Chromosomes separate into two genetically identical groups and move to opposite ends of the spindle. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media

2593: Precise development in the fruit fly embryo
2593: Precise development in the fruit fly embryo
This 2-hour-old fly embryo already has a blueprint for its formation, and the process for following it is so precise that the difference of just a few key molecules can change the plans. Here, blue marks a high concentration of Bicoid, a key signaling protein that directs the formation of the fly's head. It also regulates another important protein, Hunchback (green), that further maps the head and thorax structures and partitions the embryo in half (red is DNA). The yellow dots overlaying the embryo plot the concentration of Bicoid versus Hunchback proteins within each nucleus. The image illustrates the precision with which an embryo interprets and locates its halfway boundary, approaching limits set by simple physical principles. This image was a finalist in the 2008 Drosophila Image Award.
Thomas Gregor, Princeton University
View Media
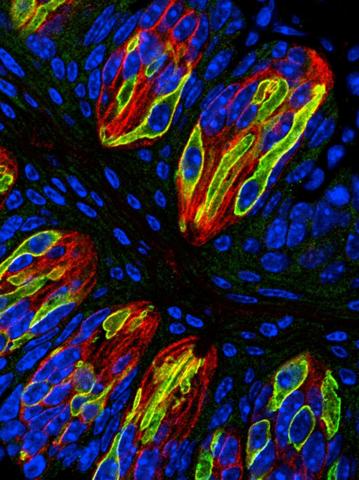
3444: Taste buds signal different tastes through ATP release
3444: Taste buds signal different tastes through ATP release
Taste buds in a mouse tongue epithelium with types I, II, and III taste cells visualized by cell-type-specific fluorescent antibodies. Type II taste bud cells signal sweet, bitter, and umami tastes to the central nervous system by releasing ATP through the voltage-gated ion channel CALHM1. Researchers used a confocal microscope to capture this image, which shows all taste buds in red, type II taste buds in green, and DNA in blue.
More information about this work can be found in the Nature letter "CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes” by Taruno et. al.
More information about this work can be found in the Nature letter "CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes” by Taruno et. al.
Aki Taruno, Perelman School of Medicine, University of Pennsylvania
View Media
2437: Hydra 01
2437: Hydra 01
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
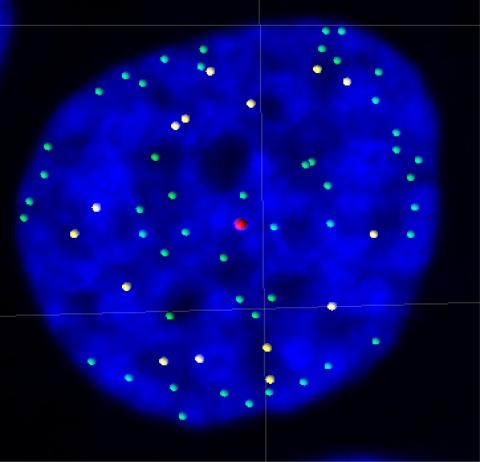
3484: Telomeres on outer edge of nucleus during cell division
3484: Telomeres on outer edge of nucleus during cell division
New research shows telomeres moving to the outer edge of the nucleus after cell division, suggesting these caps that protect chromosomes also may play a role in organizing DNA.
Laure Crabbe, Jamie Kasuboski and James Fitzpatrick, Salk Institute for Biological Studies
View Media
6521: Yeast art depicting the New York City skyline
6521: Yeast art depicting the New York City skyline
This skyline of New York City was created by “printing” nanodroplets containing yeast (Saccharomyces cerevisiae) onto a large plate. Each dot is a separate yeast colony. As the colonies grew, a picture emerged, creating art. To make the different colors shown here, yeast strains were genetically engineered to produce pigments naturally made by bacteria, fungi, and sea creatures such as coral and sea anemones. Using genes from other organisms to make biological compounds paves the way toward harnessing yeast in the production of other useful molecules, from food to fuels and drugs.
Michael Shen, Ph.D., Jasmine Temple, Leslie Mitchell, Ph.D., and Jef Boeke, Ph.D., New York University School of Medicine; and Nick Phillips, James Chuang, Ph.D., and Jiarui Wang, Johns Hopkins University.
View Media
3597: DNA replication origin recognition complex (ORC)
3597: DNA replication origin recognition complex (ORC)
A study published in March 2012 used cryo-electron microscopy to determine the structure of the DNA replication origin recognition complex (ORC), a semi-circular, protein complex (yellow) that recognizes and binds DNA to start the replication process. The ORC appears to wrap around and bend approximately 70 base pairs of double stranded DNA (red and blue). Also shown is the protein Cdc6 (green), which is also involved in the initiation of DNA replication. Related to video 3307 that shows the structure from different angles. From a Brookhaven National Laboratory news release, "Study Reveals How Protein Machinery Binds and Wraps DNA to Start Replication."
Huilin Li, Brookhaven National Laboratory
View Media
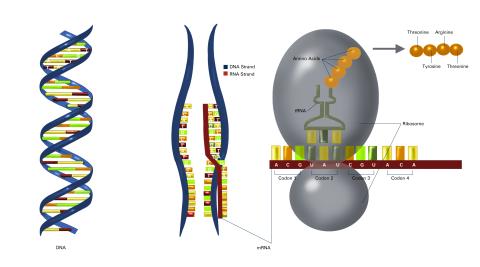
2548: Central dogma, illustrated (with labels)
2548: Central dogma, illustrated (with labels)
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2549 for a numbered version of this illustration and 2547 for an unlabeled version. Featured in The New Genetics.
Crabtree + Company
View Media

6756: Honeybees marked with paint
6756: Honeybees marked with paint
Researchers doing behavioral experiments with honeybees sometimes use paint or enamel to give individual bees distinguishing marks. The elaborate social structure and impressive learning and navigation abilities of bees make them good models for behavioral and neurobiological research. Since the sequencing of the honeybee genome, published in 2006, bees have been used increasingly for research into the molecular basis for social interaction and other complex behaviors.
Gene Robinson, University of Illinois at Urbana-Champaign.
View Media
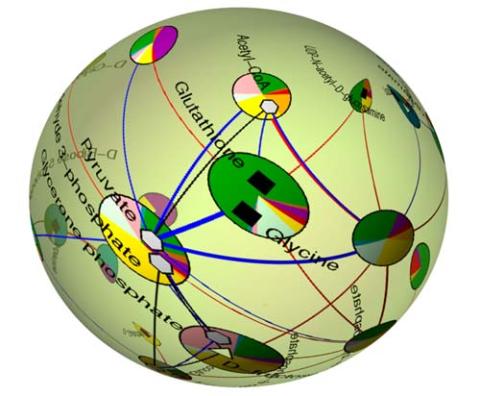
2331: Statistical cartography
2331: Statistical cartography
Like a world of its own, this sphere represents all the known chemical reactions in the E. coli bacterium. The colorful circles on the surface symbolize sets of densely interconnected reactions. The lines between the circles show additional connecting reactions. The shapes inside the circles are landmark molecules, like capital cities on a map, that either act as hubs for many groups of reactions, are highly conserved among species, or both. Molecules that connect far-flung reactions on the sphere are much more conserved during evolution than molecules that connect reactions within a single circle. This statistical cartography could reveal insights about other complex systems, such as protein interactions and gene regulation networks.
Luis A. Nunes Amaral, Northwestern University
View Media
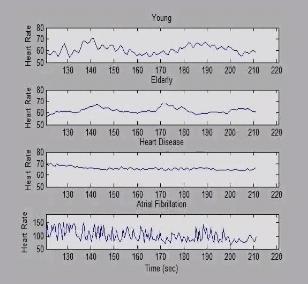
3596: Heart rates time series image
3596: Heart rates time series image
These time series show the heart rates of four different individuals. Automakers use steel scraps to build cars, construction companies repurpose tires to lay running tracks, and now scientists are reusing previously discarded medical data to better understand our complex physiology. Through a website called PhysioNet developed in part by Beth Israel Deaconess Medical Center cardiologist Ary Goldberger, scientists can access complete physiologic recordings, such as heart rate, respiration, brain activity and gait. They then can use free software to analyze the data and find patterns in it. The patterns could ultimately help health care professionals diagnose and treat health conditions like congestive heart failure, sleeping disorders, epilepsy and walking problems. PhysioNet is supported by NIH's National Institute of Biomedical Imaging and Bioengineering as well as by NIGMS.
Madalena Costa and Ary Goldberger, Beth Israel Deaconess Medical Center
View Media

5755: Autofluorescent xanthophores in zebrafish skin
5755: Autofluorescent xanthophores in zebrafish skin
Pigment cells are cells that give skin its color. In fishes and amphibians, like frogs and salamanders, pigment cells are responsible for the characteristic skin patterns that help these organisms to blend into their surroundings or attract mates. The pigment cells are derived from neural crest cells, which are cells originating from the neural tube in the early embryo. This image shows pigment cells called xanthophores in the skin of zebrafish; the cells glow (autofluoresce) brightly under light giving the fish skin a shiny, lively appearance. Investigating pigment cell formation and migration in animals helps answer important fundamental questions about the factors that control pigmentation in the skin of animals, including humans. Related to images 5754, 5756, 5757 and 5758.
David Parichy, University of Washington
View Media
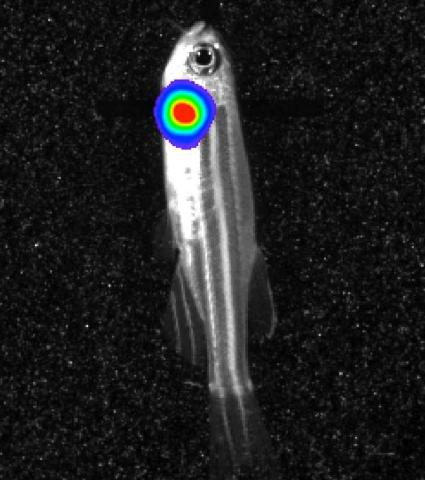
3558: Bioluminescent imaging in adult zebrafish - lateral view
3558: Bioluminescent imaging in adult zebrafish - lateral view
Luciferase-based imaging enables visualization and quantification of internal organs and transplanted cells in live adult zebrafish. In this image, a cardiac muscle-restricted promoter drives firefly luciferase expression (lateral view).
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
For imagery of both the lateral and overhead view go to 3556.
For imagery of the overhead view go to 3557.
For more information about the illumated area go to 3559.
Kenneth Poss, Duke University
View Media
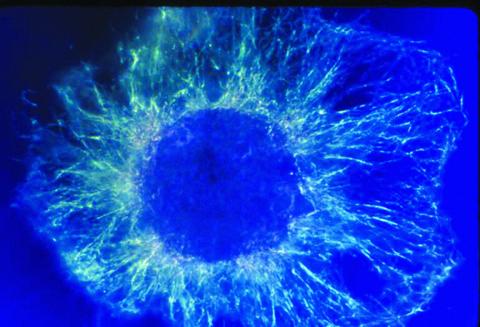
1058: Lily mitosis 01
1058: Lily mitosis 01
A light microscope image shows the chromosomes, stained dark blue, in a dividing cell of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones.
Andrew S. Bajer, University of Oregon, Eugene
View Media

3547: Master clock of the mouse brain
3547: Master clock of the mouse brain
An image of the area of the mouse brain that serves as the 'master clock,' which houses the brain's time-keeping neurons. The nuclei of the clock cells are shown in blue. A small molecule called VIP, shown in green, enables neurons in the central clock in the mammalian brain to synchronize.
Erik Herzog, Washington University in St. Louis
View Media

2395: Fungal lipase (1)
2395: Fungal lipase (1)
Crystals of fungal lipase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
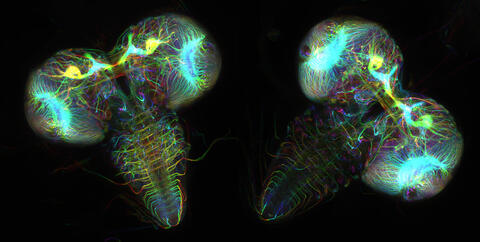
6808: Fruit fly larvae brains showing tubulin
6808: Fruit fly larvae brains showing tubulin
Two fruit fly (Drosophila melanogaster) larvae brains with neurons expressing fluorescently tagged tubulin protein. Tubulin makes up strong, hollow fibers called microtubules that play important roles in neuron growth and migration during brain development. This image was captured using confocal microscopy, and the color indicates the position of the neurons within the brain.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
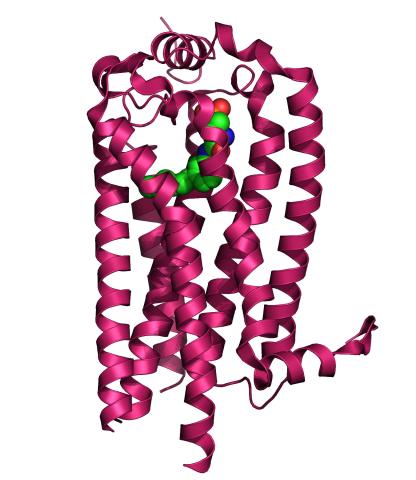
3362: Sphingolipid S1P1 receptor
3362: Sphingolipid S1P1 receptor
The receptor is shown bound to an antagonist, ML056.
Raymond Stevens, The Scripps Research Institute
View Media
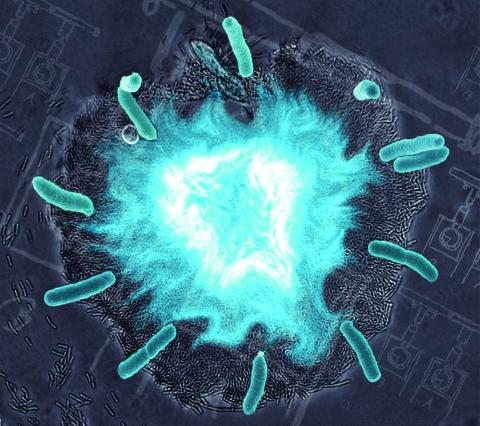
2725: Supernova bacteria
2725: Supernova bacteria
Bacteria engineered to act as genetic clocks flash in synchrony. Here, a "supernova" burst in a colony of coupled genetic clocks just after reaching critical cell density. Superimposed: A diagram from the notebook of Christiaan Huygens, who first characterized synchronized oscillators in the 17th century.
Jeff Hasty, UCSD
View Media