Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
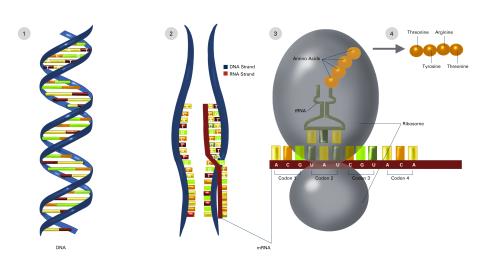
2549: Central dogma, illustrated (with labels and numbers for stages)
2549: Central dogma, illustrated (with labels and numbers for stages)
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a version of this illustration that isn't numbered and 2547 for a an entirely unlabeled version. Featured in The New Genetics.
Crabtree + Company
View Media
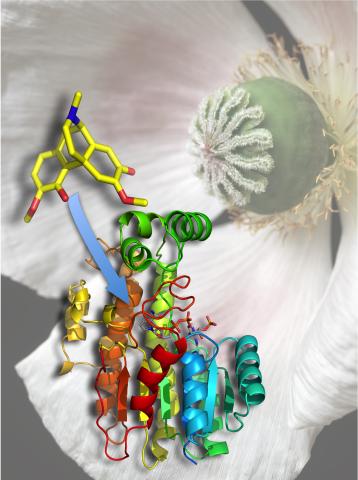
3422: Atomic Structure of Poppy Enzyme
3422: Atomic Structure of Poppy Enzyme
The atomic structure of the morphine biosynthetic enzyme salutaridine reductase bound to the cofactor NADPH. The substrate salutaridine is shown entering the active site.
Judy Coyle, Donald Danforth Plant Science Center
View Media
6792: Yeast cells with nuclei and contractile rings
6792: Yeast cells with nuclei and contractile rings
Yeast cells with nuclei shown in green and contractile rings shown in magenta. Nuclei store DNA, and contractile rings help cells divide. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6793, 6794, 6797, 6798, and videos 6795 and 6796.
Related to images 6791, 6793, 6794, 6797, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
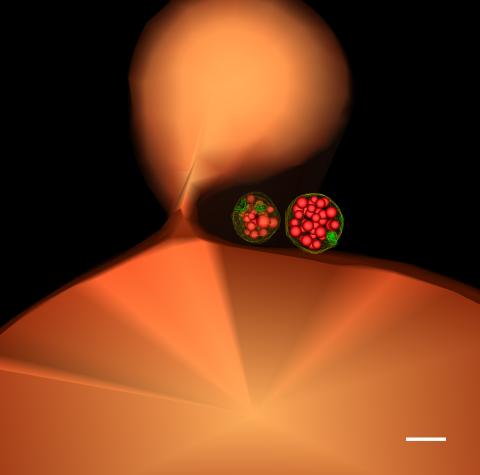
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
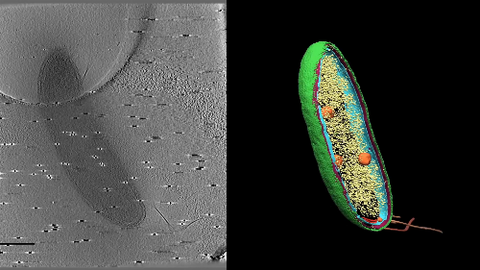
6569: Cryo-electron tomography of a Caulobacter bacterium
6569: Cryo-electron tomography of a Caulobacter bacterium
3D image of Caulobacter bacterium with various components highlighted: cell membranes (red and blue), protein shell (green), protein factories known as ribosomes (yellow), and storage granules (orange).
Peter Dahlberg, Stanford University.
View Media

6804: Staphylococcus aureus in the porous coating of a femoral hip stem
6804: Staphylococcus aureus in the porous coating of a femoral hip stem
Staphylococcus aureus bacteria (blue) on the porous coating of a femoral hip stem used in hip replacement surgery. The relatively rough surface of an implant is a favorable environment for bacteria to attach and grow. This can lead to the development of biofilms, which can cause infections. The researchers who took this image are working to understand where biofilms are likely to develop. This knowledge could support the prevention and treatment of infections. A scanning electron microscope was used to capture this image.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6803 and video 6805.
Paul Stoodley, The Ohio State University.
View Media
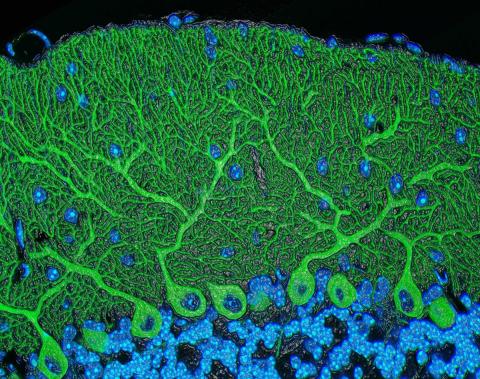
5795: Mouse cerebellum
5795: Mouse cerebellum
The cerebellum is the brain's locomotion control center. Found at the base of your brain, the cerebellum is a single layer of tissue with deep folds like an accordion. People with damage to this region of the brain often have difficulty with balance, coordination and fine motor skills.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5800.
This image of a mouse cerebellum is part of a collection of such images in different colors and at different levels of magnification from the National Center for Microscopy and Imaging Research (NCMIR). Related to image 5800.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
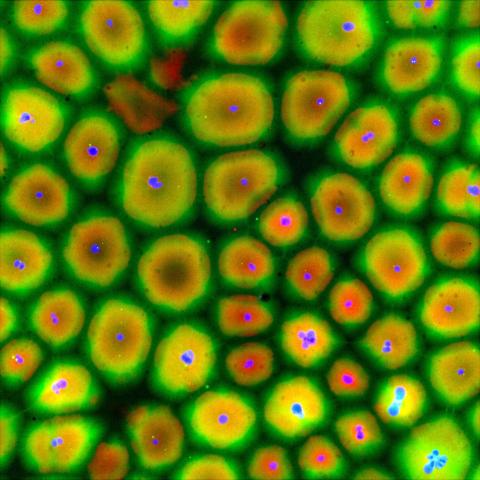
6584: Cell-like compartments from frog eggs
6584: Cell-like compartments from frog eggs
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media

2667: Glowing fish
2667: Glowing fish
Professor Marc Zimmer's family pets, including these fish, glow in the dark in response to blue light. Featured in the September 2009 issue of Findings.
View Media
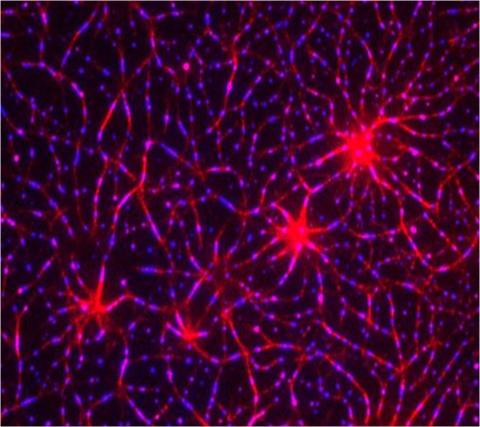
3787: In vitro assembly of a cell-signaling pathway
3787: In vitro assembly of a cell-signaling pathway
T cells are white blood cells that are important in defending the body against bacteria, viruses and other pathogens. Each T cell carries proteins, called T-cell receptors, on its surface that are activated when they come in contact with an invader. This activation sets in motion a cascade of biochemical changes inside the T cell to mount a defense against the invasion. Scientists have been interested for some time what happens after a T-cell receptor is activated. One obstacle has been to study how this signaling cascade, or pathway, proceeds inside T cells.
In this image, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The image shows two key steps during the signaling process: clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to video 3786.
In this image, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The image shows two key steps during the signaling process: clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to video 3786.
Xiaolei Su, HHMI Whitman Center of the Marine Biological Laboratory
View Media
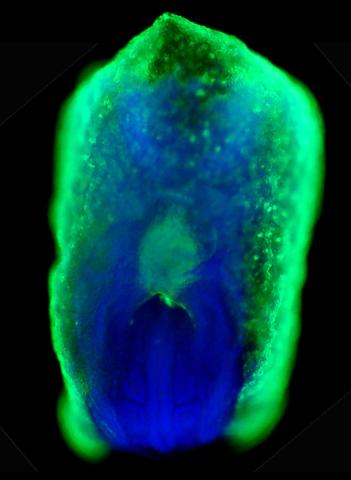
2607: Mouse embryo showing Smad4 protein
2607: Mouse embryo showing Smad4 protein
This eerily glowing blob isn't an alien or a creature from the deep sea--it's a mouse embryo just eight and a half days old. The green shell and core show a protein called Smad4. In the center, Smad4 is telling certain cells to begin forming the mouse's liver and pancreas. Researchers identified a trio of signaling pathways that help switch on Smad4-making genes, starting immature cells on the path to becoming organs. The research could help biologists learn how to grow human liver and pancreas tissue for research, drug testing and regenerative medicine. In addition to NIGMS, NIH's National Institute of Diabetes and Digestive and Kidney Diseases also supported this work.
Kenneth Zaret, Fox Chase Cancer Center
View Media

2579: Bottles of warfarin
2579: Bottles of warfarin
In 2007, the FDA modified warfarin's label to indicate that genetic makeup may affect patient response to the drug. The widely used blood thinner is sold under the brand name Coumadin®. Scientists involved in the NIH Pharmacogenetics Research Network are investigating whether genetic information can be used to improve optimal dosage prediction for patients.
Alisa Machalek, NIGMS/NIH
View Media
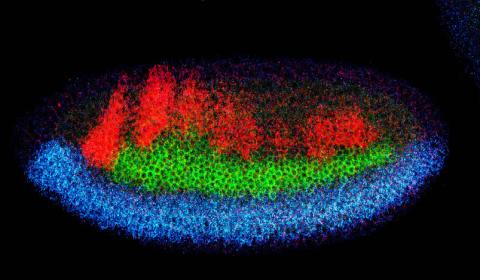
2327: Neural development
2327: Neural development
Using techniques that took 4 years to design, a team of developmental biologists showed that certain proteins can direct the subdivision of fruit fly and chicken nervous system tissue into the regions depicted here in blue, green, and red. Molecules called bone morphogenetic proteins (BMPs) helped form this fruit fly embryo. While scientists knew that BMPs play a major role earlier in embryonic development, they didn't know how the proteins help organize nervous tissue. The findings suggest that BMPs are part of an evolutionarily conserved mechanism for organizing the nervous system. The National Institute of Neurological Disorders and Stroke also supported this work.
Mieko Mizutani and Ethan Bier, University of California, San Diego, and Henk Roelink, University of Washington
View Media
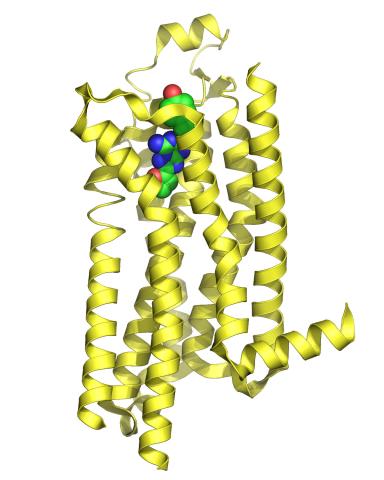
3361: A2A adenosine receptor
3361: A2A adenosine receptor
The receptor is shown bound to an inverse agonist, ZM241385.
Raymond Stevens, The Scripps Research Institute
View Media
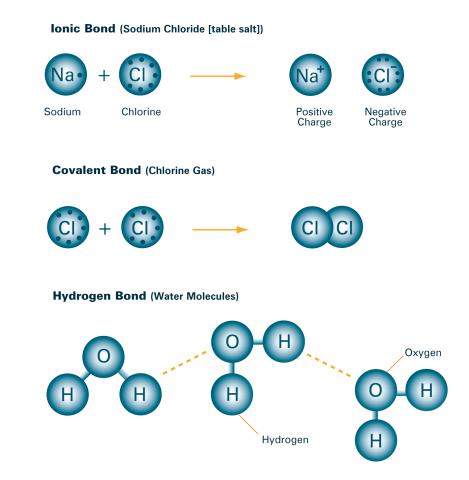
2520: Bond types (with labels)
2520: Bond types (with labels)
Ionic and covalent bonds hold molecules, like sodium chloride and chlorine gas, together. Hydrogen bonds among molecules, notably involving water, also play an important role in biology. See image 2519 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
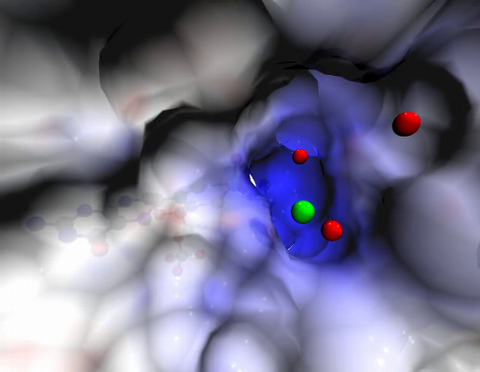
2746: Active site of sulfite oxidase
2746: Active site of sulfite oxidase
Sulfite oxidase is an enzyme that is essential for normal neurological development in children. This video shows the active site of the enzyme and its molybdenum cofactor visible as a faint ball-and-stick representation buried within the protein. The positively charged channel (blue) at the active site contains a chloride ion (green) and three water molecules (red). As the protein oscillates, one can see directly down the positively charged channel. At the bottom is the molybdenum atom of the active site (light blue) and its oxo group (red) that is transferred to sulfite to form sulfate in the catalytic reaction.
John Enemark, University of Arizona
View Media
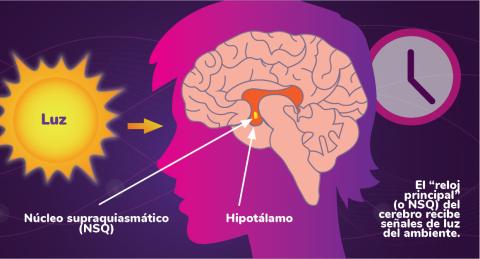
6614: Los ritmos circadianos y el núcleo supraquiasmático
6614: Los ritmos circadianos y el núcleo supraquiasmático
Los ritmos circadianos son cambios físicos, mentales y de comportamiento que siguen un ciclo de 24 horas. Los ritmos circadianos se ven influenciados por la luz y están regulados por el núcleo supraquiasmático del cerebro, a veces denominado el reloj principal.
Vea 6613 para la versión en inglés de esta infografía.
Vea 6613 para la versión en inglés de esta infografía.
NIGMS
View Media
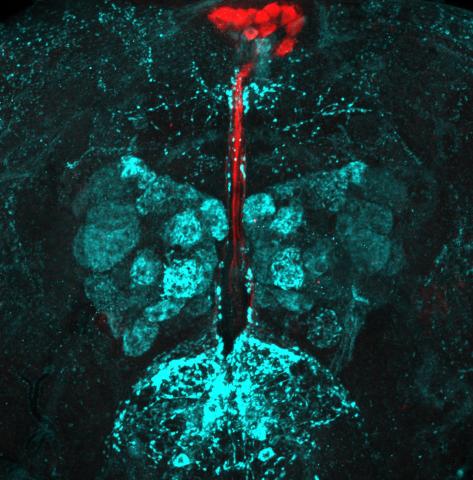
6985: Fruit fly brain responds to adipokines
6985: Fruit fly brain responds to adipokines
Drosophila adult brain showing that an adipokine (fat hormone) generates a response from neurons (aqua) and regulates insulin-producing neurons (red).
Related to images 6982, 6983, and 6984.
Related to images 6982, 6983, and 6984.
Akhila Rajan, Fred Hutchinson Cancer Center
View Media

2313: Colorful communication
2313: Colorful communication
The marine bacterium Vibrio harveyi glows when near its kind. This luminescence, which results from biochemical reactions, is part of the chemical communication used by the organisms to assess their own population size and distinguish themselves from other types of bacteria. But V. harveyi only light up when part of a large group. This communication, called quorum sensing, speaks for itself here on a lab dish, where more densely packed areas of the bacteria show up blue. Other types of bacteria use quorum sensing to release toxins, trigger disease, and evade the immune system.
Bonnie Bassler, Princeton University
View Media
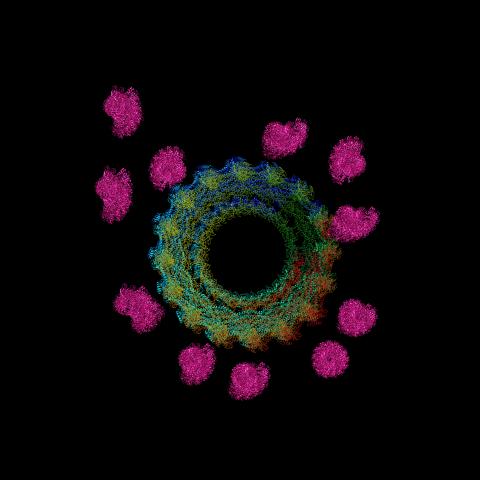
6571: Actin filaments bundled around the dynamin helical polymer
6571: Actin filaments bundled around the dynamin helical polymer
Multiple actin filaments (magenta) are organized around a dynamin helical polymer (rainbow colored) in this model derived from cryo-electron tomography. By bundling actin, dynamin increases the strength of a cell’s skeleton and plays a role in cell-cell fusion, a process involved in conception, development, and regeneration.
Elizabeth Chen, University of Texas Southwestern Medical Center.
View Media

2369: Protein purification robot in action 01
2369: Protein purification robot in action 01
A robot is transferring 96 purification columns to a vacuum manifold for subsequent purification procedures.
The Northeast Collaboratory for Structural Genomics
View Media
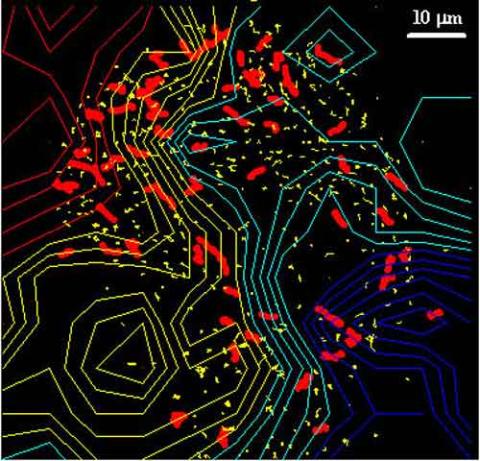
2310: Cellular traffic
2310: Cellular traffic
Like tractor-trailers on a highway, small sacs called vesicles transport substances within cells. This image tracks the motion of vesicles in a living cell. The short red and yellow marks offer information on vesicle movement. The lines spanning the image show overall traffic trends. Typically, the sacs flow from the lower right (blue) to the upper left (red) corner of the picture. Such maps help researchers follow different kinds of cellular processes as they unfold.
Alexey Sharonov and Robin Hochstrasser, University of Pennsylvania
View Media
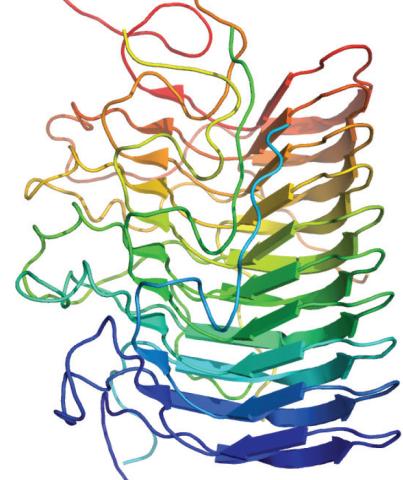
3542: Structure of amyloid-forming prion protein
3542: Structure of amyloid-forming prion protein
This structure from an amyloid-forming prion protein shows one way beta sheets can stack. Image originally appeared in a December 2012 PLOS Biology paper.
Douglas Fowler, University of Washington
View Media
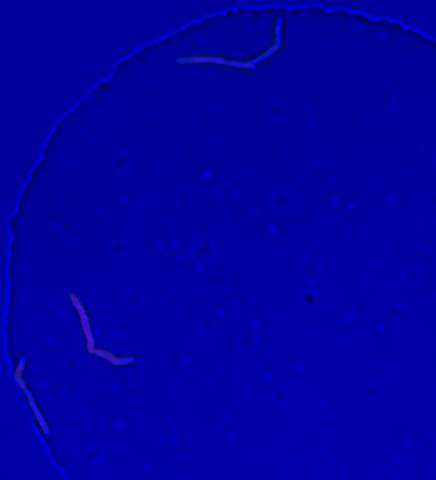
5752: Genetically identical mycobacteria respond differently to antibiotic 2
5752: Genetically identical mycobacteria respond differently to antibiotic 2
Antibiotic resistance in microbes is a serious health concern. So researchers have turned their attention to how bacteria undo the action of some antibiotics. Here, scientists set out to find the conditions that help individual bacterial cells survive in the presence of the antibiotic rifampicin. The research team used Mycobacterium smegmatis, a more harmless relative of Mycobacterium tuberculosis, which infects the lung and other organs to cause serious disease.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
In this video, genetically identical mycobacteria are growing in a miniature growth chamber called a microfluidic chamber. Using live imaging, the researchers found that individual mycobacteria will respond differently to the antibiotic, depending on the growth stage and other timing factors. The researchers used genetic tagging with green fluorescent protein to distinguish cells that can resist rifampicin and those that cannot. With this gene tag, cells tolerant of the antibiotic light up in green and those that are susceptible in violet, enabling the team to monitor the cells' responses in real time.
To learn more about how the researchers studied antibiotic resistance in mycobacteria, see this news release from Tufts University. Related to image 5751.
Bree Aldridge, Tufts University
View Media
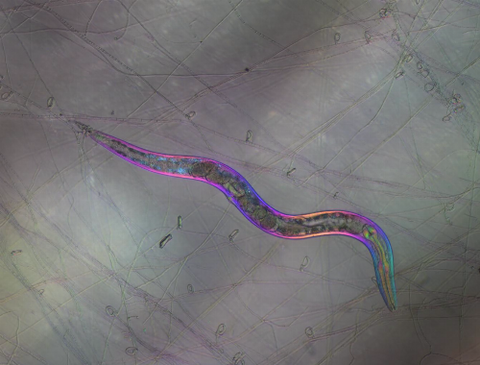
6963: C. elegans trapped by carnivorous fungus
6963: C. elegans trapped by carnivorous fungus
Real-time footage of Caenorhabditis elegans, a tiny roundworm, trapped by a carnivorous fungus, Arthrobotrys dactyloides. This fungus makes ring traps in response to the presence of C. elegans. When a worm enters a ring, the trap rapidly constricts so that the worm cannot move away, and the fungus then consumes the worm. The size of the imaged area is 0.7mm x 0.9mm.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
This video was obtained with a polychromatic polarizing microscope (PPM) in white light that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about PPM can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
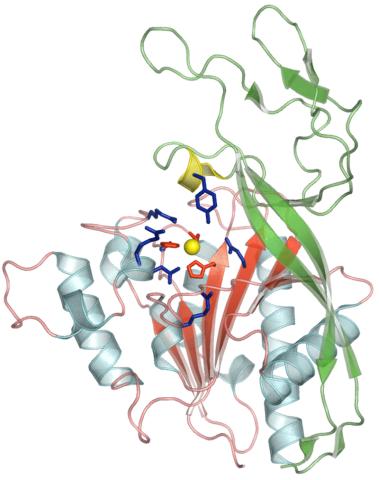
2352: Human aspartoacylase
2352: Human aspartoacylase
Model of aspartoacylase, a human enzyme involved in brain metabolism.
Center for Eukaryotic Structural Genomics, PSI
View Media
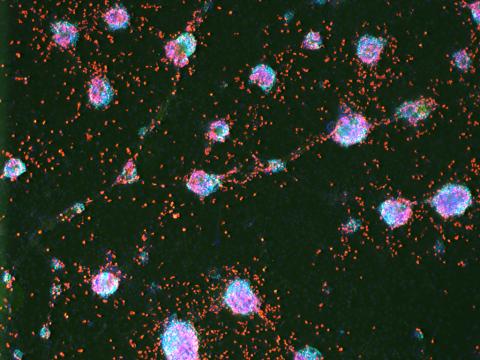
3277: Human ES cells turn into insulin-producing cells
3277: Human ES cells turn into insulin-producing cells
Human embryonic stem cells were differentiated into cells like those found in the pancreas (blue), which give rise to insulin-producing cells (red). When implanted in mice, the stem cell-derived pancreatic cells can replace the insulin that isn't produced in type 1 diabetes. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Eugene Brandon, ViaCyte, via CIRM
View Media
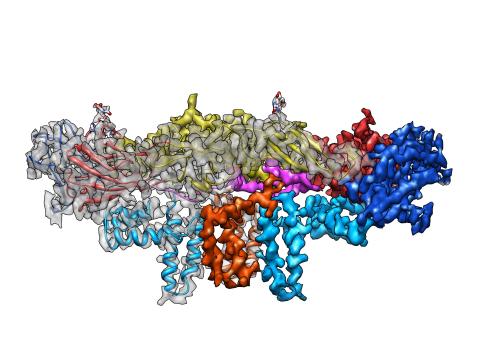
3758: Dengue virus membrane protein structure
3758: Dengue virus membrane protein structure
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. The image shows a side view of the structure of a protein composed of two smaller proteins, called E and M. Each E and M contributes two molecules to the overall protein structure (called a heterotetramer), which is important for assembling and holding together the viral membrane, i.e., the shell that surrounds the genetic material of the dengue virus. The dengue protein's structure has revealed some portions in the protein that might be good targets for developing medications that could be used to combat dengue virus infections. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. You can watch a rotating view of the dengue virus surface structure in video 3748.
Hong Zhou, UCLA
View Media
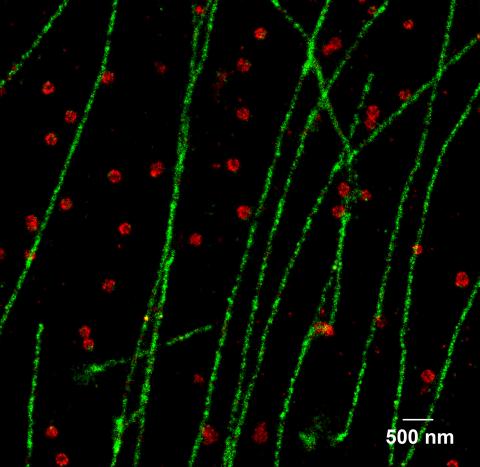
2325: Multicolor STORM
2325: Multicolor STORM
In 2006, scientists developed an optical microscopy technique enabling them to clearly see individual molecules within cells. In 2007, they took the technique, abbreviated STORM, a step further. They identified multicolored probes that let them peer into cells and clearly see multiple cellular components at the same time, such as these microtubules (green) and small hollows called clathrin-coated pits (red). Unlike conventional methods, the multicolor STORM technique produces a crisp and high resolution picture. A sharper view of how cellular components interact will likely help scientists answer some longstanding questions about cell biology.
Xiaowei Zhuang, Harvard University
View Media
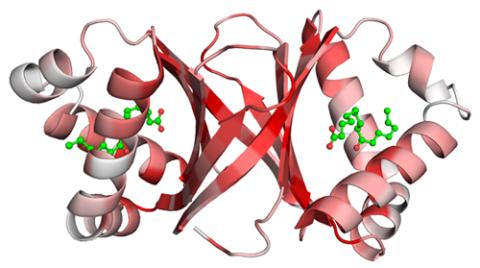
2340: Dimeric ferredoxin-like protein from an unidentified marine microbe
2340: Dimeric ferredoxin-like protein from an unidentified marine microbe
This is the first structure of a protein derived from the metagenomic sequences collected during the Sorcerer II Global Ocean Sampling project. The crystal structure shows a barrel protein with a ferredoxin-like fold and a long chain fatty acid in a deep cleft (shaded red). Featured as one of the August 2007 Protein Structure Initiative Structures of the Month.
Joint Center for Structural Genomics
View Media
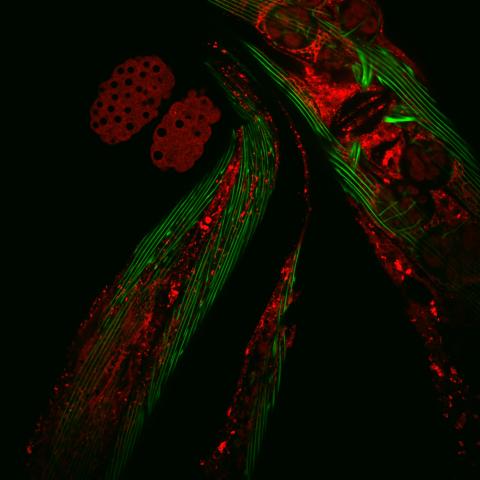
6583: Closeup of fluorescent C. elegans showing muscle and ribosomal protein
6583: Closeup of fluorescent C. elegans showing muscle and ribosomal protein
Closeup of C. elegans, tiny roundworms, with a ribosomal protein glowing red and muscle fibers glowing green. Researchers used these worms to study a molecular pathway that affects aging. The ribosomal protein is involved in protein translation and may play a role in dietary restriction-induced longevity. Image created using confocal microscopy.
View single roundworm here 6581.
View group of roundworms here 6582.
View single roundworm here 6581.
View group of roundworms here 6582.
Jarod Rollins, Mount Desert Island Biological Laboratory.
View Media
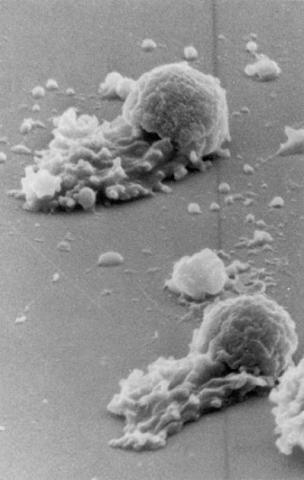
3489: Worm sperm
3489: Worm sperm
To develop a system for studying cell motility in unnatrual conditions -- a microscope slide instead of the body -- Tom Roberts and Katsuya Shimabukuro at Florida State University disassembled and reconstituted the motility parts used by worm sperm cells.
Tom Roberts, Florida State University
View Media
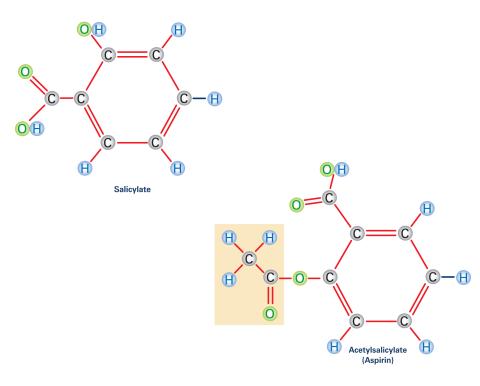
2530: Aspirin (with labels)
2530: Aspirin (with labels)
Acetylsalicylate (bottom) is the aspirin of today. Adding a chemical tag called an acetyl group (shaded box, bottom) to a molecule derived from willow bark (salicylate, top) makes the molecule less acidic (and easier on the lining of the digestive tract), but still effective at relieving pain. See image 2529 for an unlabeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
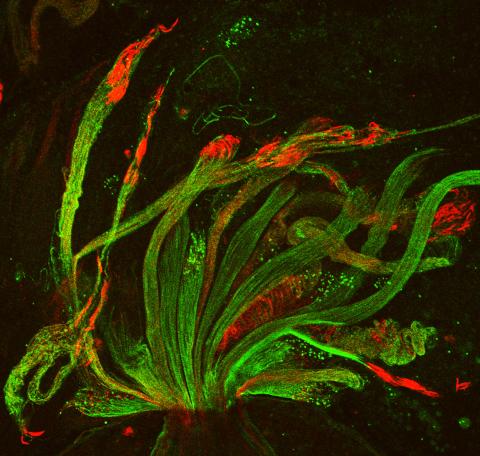
3590: Fruit fly spermatids
3590: Fruit fly spermatids
Developing spermatids (precursors of mature sperm cells) begin as small, round cells and mature into long-tailed, tadpole-shaped ones. In the sperm cell's head is the cell nucleus; in its tail is the power to outswim thousands of competitors to fertilize an egg. As seen in this microscopy image, fruit fly spermatids start out as groups of interconnected cells. A small lipid molecule called PIP2 helps spermatids tell their heads from their tails. Here, PIP2 (red) marks the nuclei and a cell skeleton-building protein called tubulin (green) marks the tails. When PIP2 levels are too low, some spermatids get mixed up and grow with their heads at the wrong end. Because sperm development is similar across species, studies in fruit flies could help researchers understand male infertility in humans.
Lacramioara Fabian, The Hospital for Sick Children, Toronto, Canada
View Media
6589: Cell-like compartments emerging from scrambled frog eggs 3
6589: Cell-like compartments emerging from scrambled frog eggs 3
Cell-like compartments spontaneously emerge from scrambled frog eggs. Endoplasmic reticulum (red) and microtubules (green) are visible. Video created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6584, 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
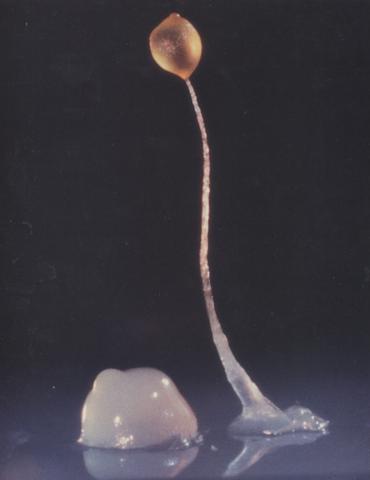
2684: Dicty fruit
2684: Dicty fruit
Dictyostelium discoideum is a microscopic amoeba. A group of 100,000 form a mound as big as a grain of sand. Featured in The New Genetics.
View Media

6886: Neutrophil-like cells migrating in a microfluidic chip
6886: Neutrophil-like cells migrating in a microfluidic chip
Neutrophil-like cells (blue) in a microfluidic chip preferentially migrating toward LTB4 over fMLP. A neutrophil is a type of white blood cell that is part of the immune system and helps the body fight infection. Both LTB4 and fMLP are molecules involved in immune response. Microfluidic chips are small devices containing microscopic channels, and they are used in a range of applications, from basic research on cells to pathogen detection. The scale bar in this video is 500μm.
Caroline Jones, University of Texas at Dallas.
View Media
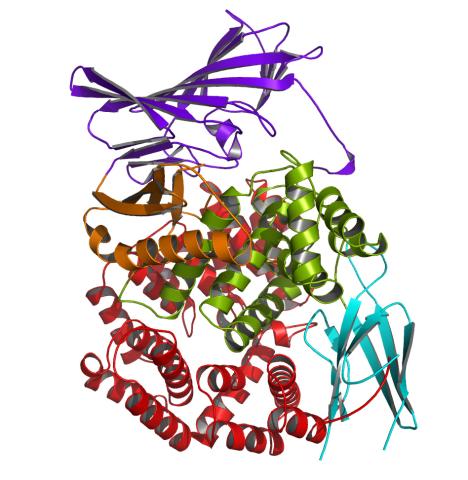
2341: Aminopeptidase N from N. meningitidis
2341: Aminopeptidase N from N. meningitidis
Model of the enzyme aminopeptidase N from the human pathogen Neisseria meningitidis, which can cause meningitis epidemics. The structure provides insight on the active site of this important molecule.
Midwest Center for Structural Genomics, PSI
View Media
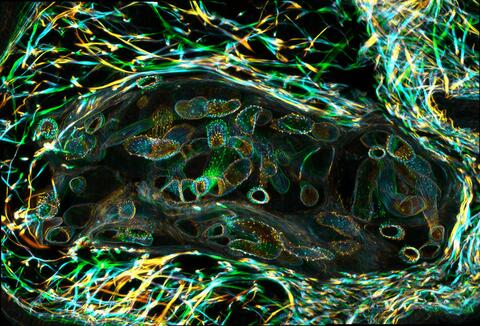
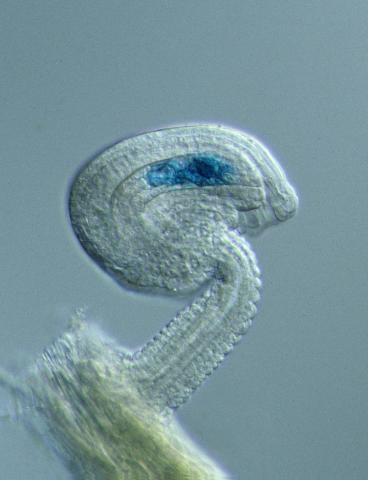
3627: Larvae from the parasitic worm that causes schistosomiasis
3627: Larvae from the parasitic worm that causes schistosomiasis
The parasitic worm that causes schistosomiasis hatches in water and grows up in a freshwater snail, as shown here. Once mature, the worm swims back into the water, where it can infect people through skin contact. Initially, an infected person might have a rash, itchy skin, or flu-like symptoms, but the real damage is done over time to internal organs.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Bo Wang and Phillip A. Newmark, University of Illinois at Urbana-Champaign, 2013 FASEB BioArt winner
View Media
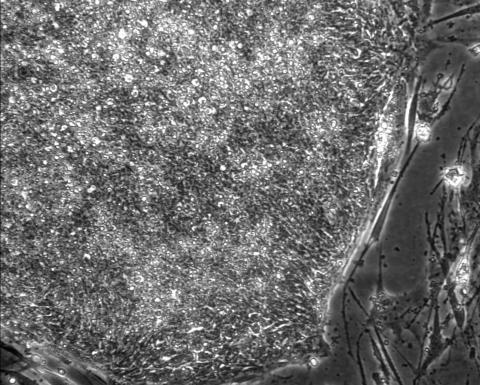
2603: Induced stem cells from adult skin 01
2603: Induced stem cells from adult skin 01
These cells are induced stem cells made from human adult skin cells that were genetically reprogrammed to mimic embryonic stem cells. The induced stem cells were made potentially safer by removing the introduced genes and the viral vector used to ferry genes into the cells, a loop of DNA called a plasmid. The work was accomplished by geneticist Junying Yu in the laboratory of James Thomson, a University of Wisconsin-Madison School of Medicine and Public Health professor and the director of regenerative biology for the Morgridge Institute for Research.
James Thomson, University of Wisconsin-Madison
View Media
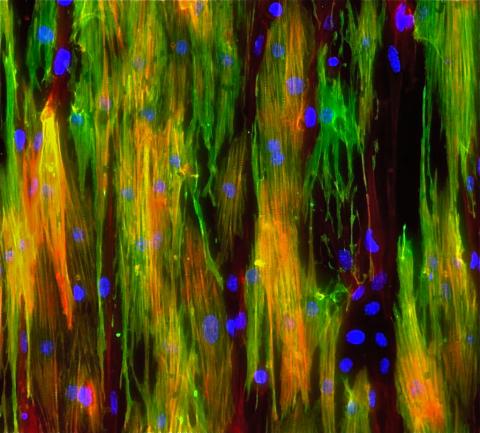
3283: Mouse heart muscle cells 02
3283: Mouse heart muscle cells 02
This image shows neonatal mouse heart cells. These cells were grown in the lab on a chip that aligns the cells in a way that mimics what is normally seen in the body. Green shows the muscle protein toponin I. Red indicates the muscle protein actin, and blue indicates the cell nuclei. The work shown here was part of a study attempting to grow heart tissue in the lab to repair damage after a heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to images 3281 and 3282.
Kara McCloskey lab, University of California, Merced, via CIRM
View Media
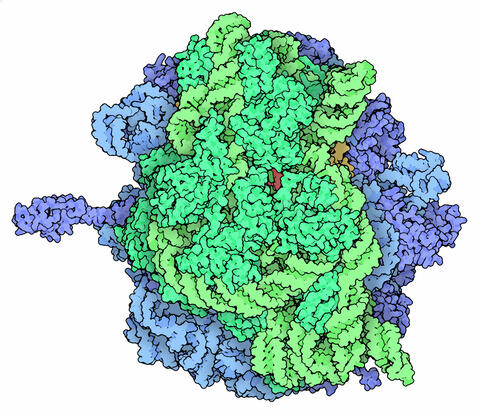
5780: Ribosome illustration from PDB
5780: Ribosome illustration from PDB
Ribosomes are complex machines made up of more than 50 proteins and three or four strands of genetic material called ribosomal RNA (rRNA). The busy cellular machines make proteins, which are critical to almost every structure and function in the cell. To do so, they read protein-building instructions, which come as strands of messenger RNA. Ribosomes are found in all forms of cellular life—people, plants, animals, even bacteria. This illustration of a bacterial ribosome was produced using detailed information about the position of every atom in the complex. Several antibiotic medicines work by disrupting bacterial ribosomes but leaving human ribosomes alone. Scientists are carefully comparing human and bacterial ribosomes to spot differences between the two. Structures that are present only in the bacterial version could serve as targets for new antibiotic medications.
From PDB’s Molecule of the Month collection (direct link: http://pdb101.rcsb.org/motm/121) Molecule of the Month illustrations are available under a CC-BY-4.0 license. Attribution should be given to David S. Goodsell and the RCSB PDB.
View Media
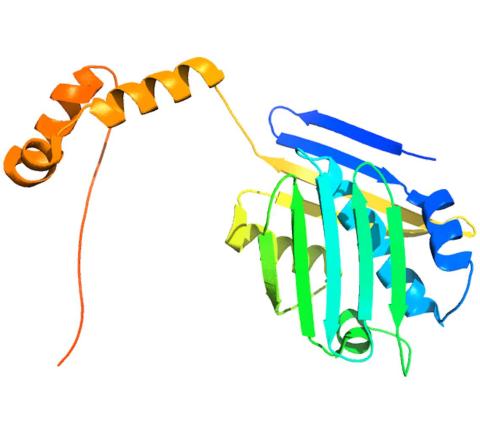
3355: Hsp33 figure 2
3355: Hsp33 figure 2
Featured in the March 15, 2012 issue of Biomedical Beat. Related to Hsp33 Figure 1, image 3354.
Ursula Jakob and Dana Reichmann, University of Michigan
View Media
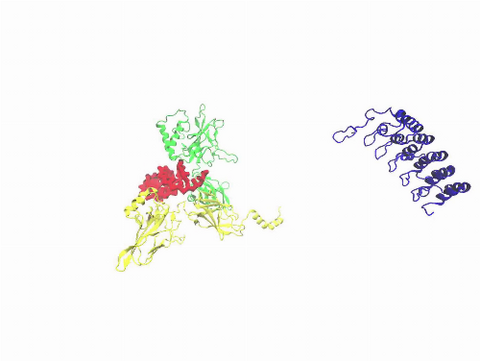
3729: A molecular switch strips transcription factor from DNA
3729: A molecular switch strips transcription factor from DNA
In this video, Rice University scientists used molecular modeling with a mathematical algorithm called AWSEM (for associative memory, water-mediated, structure and energy model) and structural data to analyze how a transcription factor called nuclear factor kappa B (NFkB) is removed from DNA to stop gene activation. AWSEM uses the interacting energies of their components to predict how proteins fold. At the start, the NFkB dimer (green and yellow, in the center) grips DNA (red, to the left), which activates the transcription of genes. IkB (blue, to the right), an inhibitor protein, stops transcription when it binds to NFkB and forces the dimer to twist and release its hold on DNA. The yellow domain at the bottom of IkB is the PEST domain, which binds first to NFkB. For more details about this mechanism called molecular stripping, see here.
Davit Potoyan and Peter Wolynes
View Media
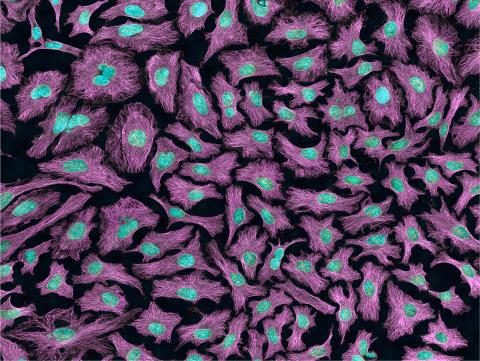
3520: HeLa cells
3520: HeLa cells
Multiphoton fluorescence image of HeLa cells with cytoskeletal microtubules (magenta) and DNA (cyan). Nikon RTS2000MP custom laser scanning microscope. See related images 3518, 3519, 3521, 3522.
National Center for Microscopy and Imaging Research (NCMIR)
View Media
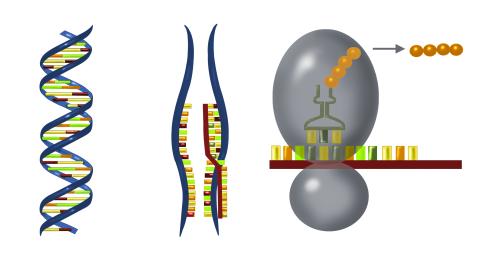
2547: Central dogma, illustrated
2547: Central dogma, illustrated
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a labeled version of this illustration and 2549 for a labeled and numbered version. Featured in The New Genetics.
Crabtree + Company
View Media
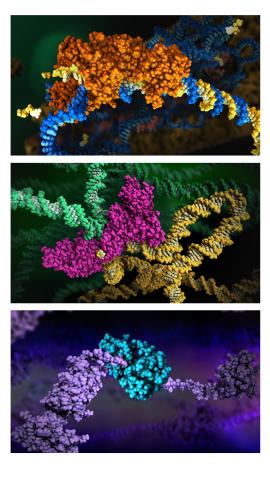
6999: HIV enzyme
6999: HIV enzyme
These images model the molecular structures of three enzymes with critical roles in the life cycle of the human immunodeficiency virus (HIV). At the top, reverse transcriptase (orange) creates a DNA copy (yellow) of the virus's RNA genome (blue). In the middle image, integrase (magenta) inserts this DNA copy in the DNA genome (green) of the infected cell. At the bottom, much later in the viral life cycle, protease (turquoise) chops up a chain of HIV structural protein (purple) to generate the building blocks for making new viruses. See these enzymes in action on PDB 101’s video A Molecular View of HIV Therapy.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
3286: Retinal pigment epithelium derived from human ES cells
3286: Retinal pigment epithelium derived from human ES cells
This color-enhanced image is a scanning electron microscope image of retinal pigment epithelial (RPE) cells derived from human embryonic stem cells. The cells are remarkably similar to normal RPE cells, growing in a hexagonal shape in a single, well-defined layer. This kind of retinal cell is responsible for macular degeneration, the most common cause of blindness. Image and caption information courtesy of the California Institute for Regenerative Medicine. Related to image 3287.
David Hinton lab, University of Southern California, via CIRM
View Media
2418: Genetic imprinting in Arabidopsis
2418: Genetic imprinting in Arabidopsis
This delicate, birdlike projection is an immature seed of the Arabidopsis plant. The part in blue shows the cell that gives rise to the endosperm, the tissue that nourishes the embryo. The cell is expressing only the maternal copy of a gene called MEDEA. This phenomenon, in which the activity of a gene can depend on the parent that contributed it, is called genetic imprinting. In Arabidopsis, the maternal copy of MEDEA makes a protein that keeps the paternal copy silent and reduces the size of the endosperm. In flowering plants and mammals, this sort of genetic imprinting is thought to be a way for the mother to protect herself by limiting the resources she gives to any one embryo. Featured in the May 16, 2006, issue of Biomedical Beat.
Robert Fischer, University of California, Berkeley
View Media