Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
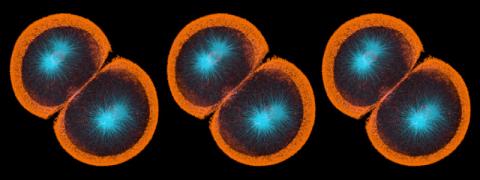
6793: Yeast cells with endocytic actin patches
6793: Yeast cells with endocytic actin patches
Yeast cells with endocytic actin patches (green). These patches help cells take in outside material. When a cell is in interphase, patches concentrate at its ends. During later stages of cell division, patches move to where the cell splits. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6794, 6797, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
3391: Protein folding video
3391: Protein folding video
Proteins are long chains of amino acids. Each protein has a unique amino acid sequence. It is still a mystery how a protein folds into the proper shape based on its sequence. Scientists hope that one day they can "watch" this folding process for any given protein. The dream has been realized, at least partially, through the use of computer simulation.
Theoretical and Computational Biophysics Group
View Media
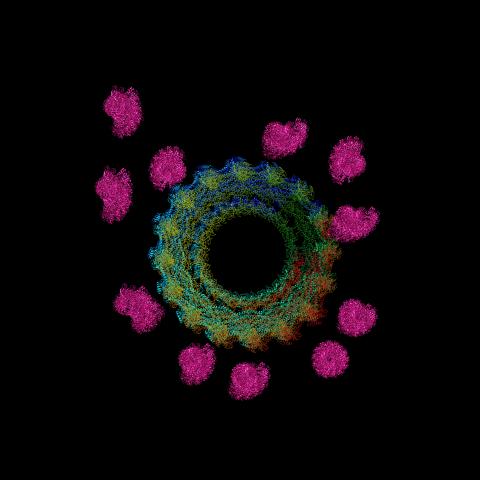
6571: Actin filaments bundled around the dynamin helical polymer
6571: Actin filaments bundled around the dynamin helical polymer
Multiple actin filaments (magenta) are organized around a dynamin helical polymer (rainbow colored) in this model derived from cryo-electron tomography. By bundling actin, dynamin increases the strength of a cell’s skeleton and plays a role in cell-cell fusion, a process involved in conception, development, and regeneration.
Elizabeth Chen, University of Texas Southwestern Medical Center.
View Media
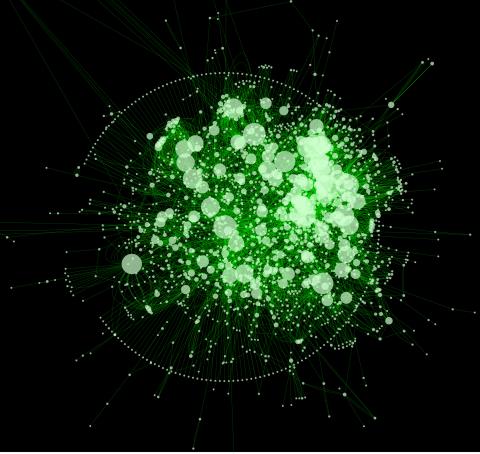
2737: Cytoscape network diagram 1
2737: Cytoscape network diagram 1
Molecular biologists are increasingly relying on bioinformatics software to visualize molecular interaction networks and to integrate these networks with data such as gene expression profiles. Related to 2749.
Keiichiro Ono, Trey Ideker lab, University of California, San Diego
View Media
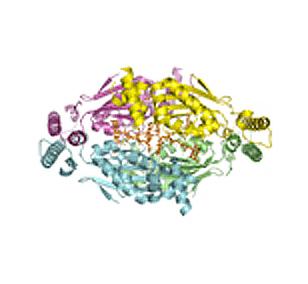
2387: Thymidylate synthase complementing protein from Thermotoga maritime
2387: Thymidylate synthase complementing protein from Thermotoga maritime
A model of thymidylate synthase complementing protein from Thermotoga maritime.
Joint Center for Structural Genomics, PSI
View Media
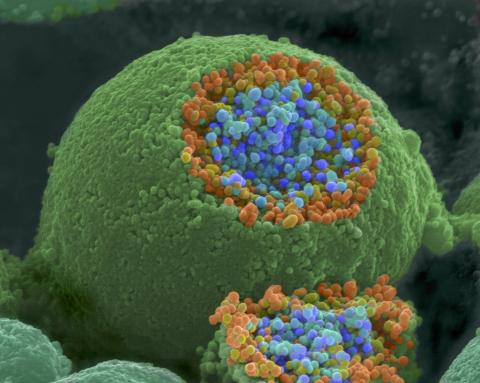
1244: Nerve ending
1244: Nerve ending
A scanning electron microscope picture of a nerve ending. It has been broken open to reveal vesicles (orange and blue) containing chemicals used to pass messages in the nervous system.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media

1283: Vesicle traffic
1283: Vesicle traffic
This illustration shows vesicle traffic inside a cell. The double membrane that bounds the nucleus flows into the ribosome-studded rough endoplasmic reticulum (purple), where membrane-embedded proteins are manufactured. Proteins are processed and lipids are manufactured in the smooth endoplasmic reticulum (blue) and Golgi apparatus (green). Vesicles that fuse with the cell membrane release their contents outside the cell. The cell can also take in material from outside by having vesicles pinch off from the cell membrane.
Judith Stoffer
View Media
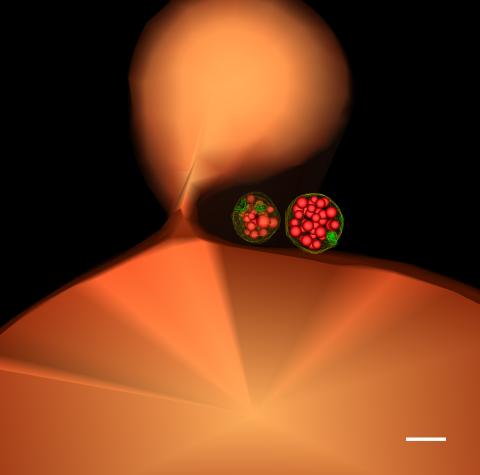
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
5769: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 1
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-in version 5767 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
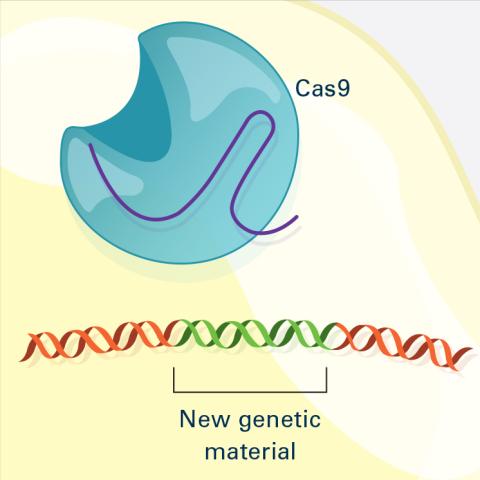
6488: CRISPR Illustration Frame 4
6488: CRISPR Illustration Frame 4
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). This frame (4 out of 4) shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media

2531: Drugs enter skin
2531: Drugs enter skin
Drugs enter different layers of skin via intramuscular, subcutaneous, or transdermal delivery methods. See image 2532 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media

3567: RSV-Infected Cell
3567: RSV-Infected Cell
Viral RNA (red) in an RSV-infected cell. More information about the research behind this image can be found in a Biomedical Beat Blog posting from January 2014.
Eric Alonas and Philip Santangelo, Georgia Institute of Technology and Emory University
View Media
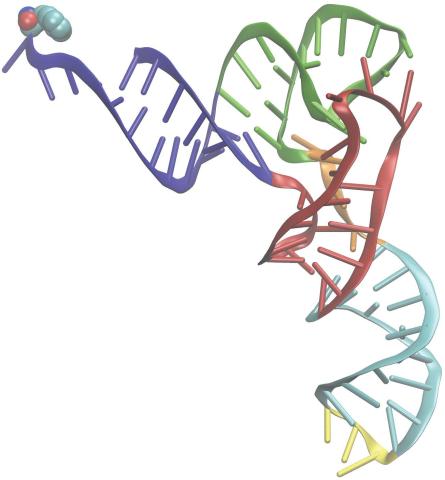
3406: Phenylalanine tRNA molecule
3406: Phenylalanine tRNA molecule
Phenylalanine tRNA showing the anticodon (yellow) and the amino acid, phenylalanine (blue and red spheres).
Patrick O'Donoghue and Dieter Soll, Yale University
View Media
6776: Tracking cells in a gastrulating zebrafish embryo
6776: Tracking cells in a gastrulating zebrafish embryo
During development, a zebrafish embryo is transformed from a ball of cells into a recognizable body plan by sweeping convergence and extension cell movements. This process is called gastrulation. Each line in this video represents the movement of a single zebrafish embryo cell over the course of 3 hours. The video was created using time-lapse confocal microscopy. Related to image 6775.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media
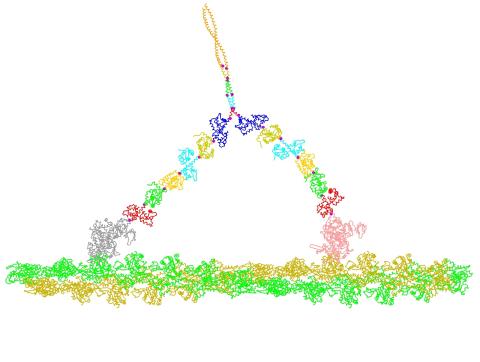
2754: Myosin V binding to actin
2754: Myosin V binding to actin
This simulation of myosin V binding to actin was created using the software tool Protein Mechanica. With Protein Mechanica, researchers can construct models using information from a variety of sources: crystallography, cryo-EM, secondary structure descriptions, as well as user-defined solid shapes, such as spheres and cylinders. The goal is to enable experimentalists to quickly and easily simulate how different parts of a molecule interact.
Simbios, NIH Center for Biomedical Computation at Stanford
View Media
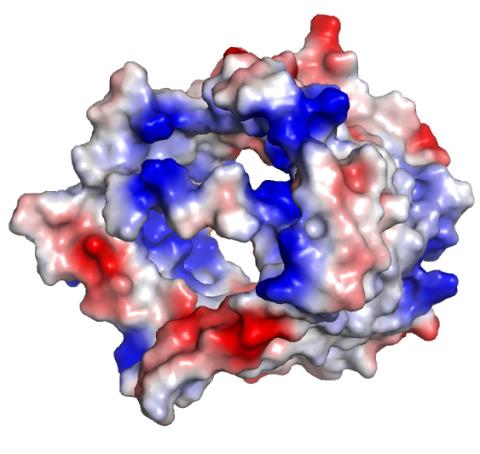
2494: VDAC-1 (3)
2494: VDAC-1 (3)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2495, and 2488.
Related to images 2491, 2495, and 2488.
Gerhard Wagner, Harvard Medical School
View Media
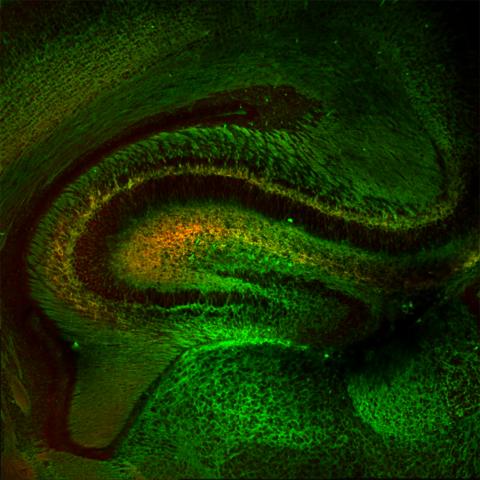
5886: Mouse Brain Cross Section
5886: Mouse Brain Cross Section
The brain sections are treated with fluorescent antibodies specific to a particular protein and visualized using serial electron microscopy (SEM).
Anton Maximov, The Scripps Research Institute, La Jolla, CA
View Media
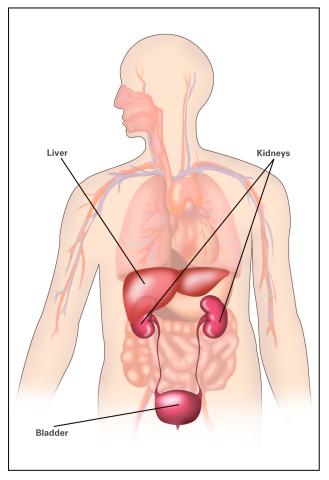
2497: Body toxins (with labels)
2497: Body toxins (with labels)
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2496 for an unlabeled version of this illustration.
Crabtree + Company
View Media
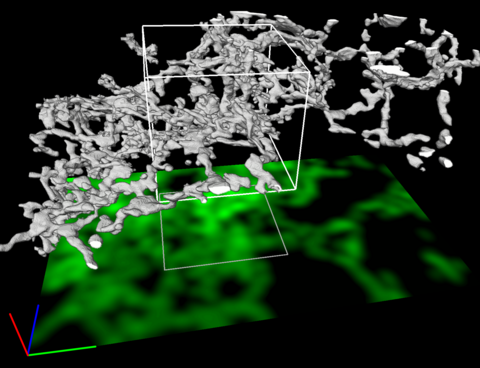
5856: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 2
5856: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 2
Three-dimensional reconstruction of a tubular matrix in a thin section of the peripheral endoplasmic reticulum between the plasma membranes of the cell. The endoplasmic reticulum (ER) is a continuous membrane that extends like a net from the envelope of the nucleus outward to the cell membrane. The ER plays several roles within the cell, such as in protein and lipid synthesis and transport of materials between organelles. Shown here are super-resolution microscopic images of the peripheral ER showing the structure of an ER tubular matrix between the plasma membranes of the cell. See image 5857 for a more detailed view of the area outlined in white in this image. For another view of the ER tubular matrix see image 5855
Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia
View Media
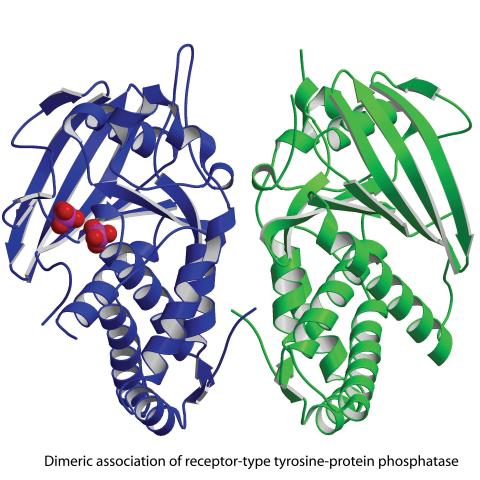
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
2349: Dimeric association of receptor-type tyrosine-protein phosphatase
Model of the catalytic portion of an enzyme, receptor-type tyrosine-protein phosphatase from humans. The enzyme consists of two identical protein subunits, shown in blue and green. The groups made up of purple and red balls represent phosphate groups, chemical groups that can influence enzyme activity. This phosphatase removes phosphate groups from the enzyme tyrosine kinase, counteracting its effects.
New York Structural GenomiX Research Consortium, PSI
View Media
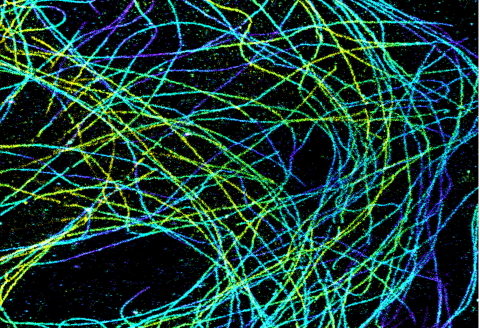
6891: Microtubules in African green monkey cells
6891: Microtubules in African green monkey cells
Microtubules in African green monkey cells. Microtubules are strong, hollow fibers that provide cells with structural support. Here, the microtubules have been color-coded based on their distance from the microscope lens: purple is closest to the lens, and yellow is farthest away. This image was captured using Stochastic Optical Reconstruction Microscopy (STORM).
Related to images 6889, 6890, and 6892.
Related to images 6889, 6890, and 6892.
Melike Lakadamyali, Perelman School of Medicine at the University of Pennsylvania.
View Media
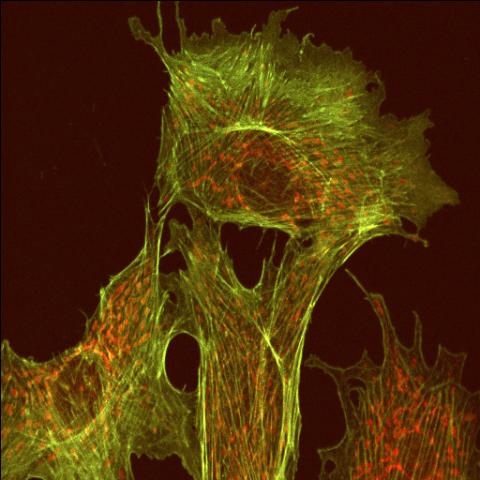
1102: Endothelial cell
1102: Endothelial cell
This image shows two components of the cytoskeleton, microtubules (green) and actin filaments (red), in an endothelial cell derived from a cow lung. The cystoskeleton provides the cell with an inner framework and enables it to move and change shape.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media
2442: Hydra 06
2442: Hydra 06
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media
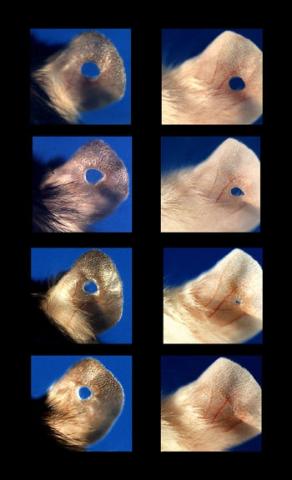
3426: Regeneration of Mouse Ears
3426: Regeneration of Mouse Ears
Normal mice, like the B6 breed pictured on the left, develop scars when their ears are pierced. The Murphy Roths Large (MRL) mice pictured on the right can grow back lost ear tissue thanks to an inactive version of the p21 gene. When researchers knocked out that same gene in other mouse breeds, their ears also healed completely without scarring. Journal Article: Clark, L.D., Clark, R.K. and Heber-Katz, E. 1998. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol 88: 35-45.
Ellen Heber-Katz, The Wistar Institute
View Media
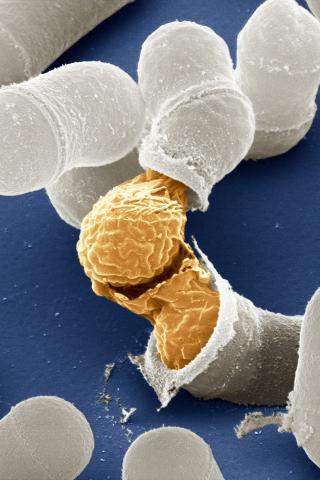
3614: Birth of a yeast cell
3614: Birth of a yeast cell
Yeast make bread, beer, and wine. And like us, yeast can reproduce sexually. A mother and father cell fuse and create one large cell that contains four offspring. When environmental conditions are favorable, the offspring are released, as shown here. Yeast are also a popular study subject for scientists. Research on yeast has yielded vast knowledge about basic cellular and molecular biology as well as about myriad human diseases, including colon cancer and various metabolic disorders.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Juergen Berger, Max Planck Institute for Developmental Biology, and Maria Langegger, Friedrich Miescher Laboratory of the Max Planck Society, Germany
View Media
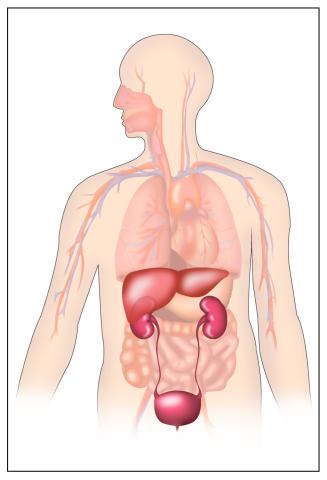
2496: Body toxins
2496: Body toxins
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2497 for a labeled version of this illustration.
Crabtree + Company
View Media

6769: Culex quinquefasciatus mosquito larva
6769: Culex quinquefasciatus mosquito larva
A mosquito larva with genes edited by CRISPR. The red-orange glow is a fluorescent protein used to track the edits. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this image developed a gene-editing toolkit for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to image 6770 and video 6771.
Valentino Gantz, University of California, San Diego.
View Media
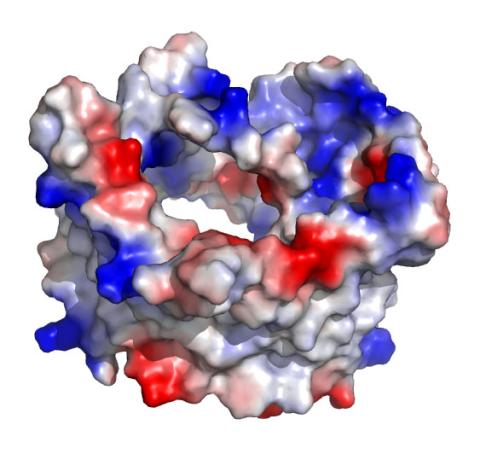
2488: VDAC-1 (1)
2488: VDAC-1 (1)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2491, 2494, and 2495.
Related to images 2491, 2494, and 2495.
Gerhard Wagner, Harvard Medical School
View Media
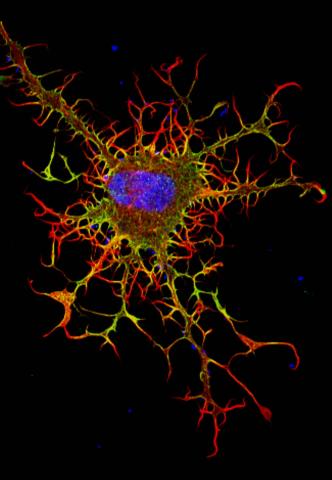
3613: Abnormal, spiky fibroblast
3613: Abnormal, spiky fibroblast
This is a fibroblast, a connective tissue cell that plays an important role in wound healing. Normal fibroblasts have smooth edges. In contrast, this spiky cell is missing a protein that is necessary for proper construction of the cell's skeleton. Its jagged shape makes it impossible for the cell to move normally. In addition to compromising wound healing, abnormal cell movement can lead to birth defects, faulty immune function, and other health problems.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Praveen Suraneni, Stowers Institute for Medical Research, Kansas City, Mo.
View Media
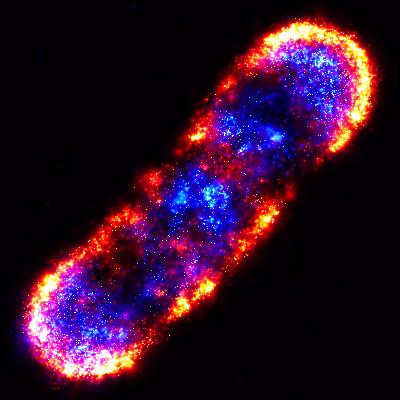
2771: Self-organizing proteins
2771: Self-organizing proteins
Under the microscope, an E. coli cell lights up like a fireball. Each bright dot marks a surface protein that tells the bacteria to move toward or away from nearby food and toxins. Using a new imaging technique, researchers can map the proteins one at a time and combine them into a single image. This lets them study patterns within and among protein clusters in bacterial cells, which don't have nuclei or organelles like plant and animal cells. Seeing how the proteins arrange themselves should help researchers better understand how cell signaling works.
View Media
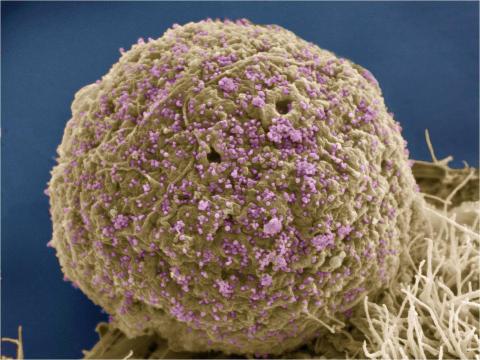
3386: HIV Infected Cell
3386: HIV Infected Cell
The human immunodeficiency virus (HIV), shown here as tiny purple spheres, causes the disease known as AIDS (for acquired immunodeficiency syndrome). HIV can infect multiple cells in your body, including brain cells, but its main target is a cell in the immune system called the CD4 lymphocyte (also called a T-cell or CD4 cell).
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
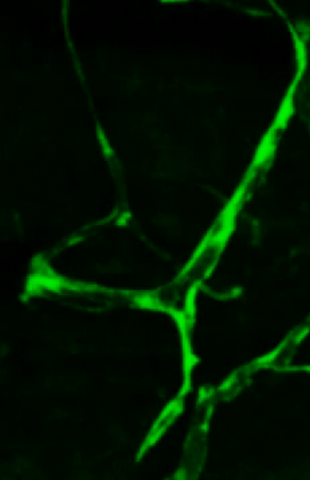
3405: Disrupted and restored vasculature development in frog embryos
3405: Disrupted and restored vasculature development in frog embryos
Disassembly of vasculature and reassembly after addition and then washout of 250 µM TBZ in kdr:GFP frogs. Related to images 3403 and 3404.
Hye Ji Cha, University of Texas at Austin
View Media
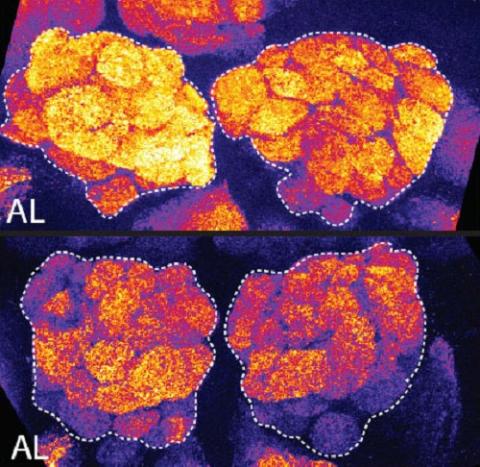
3490: Brains of sleep-deprived and well-rested fruit flies
3490: Brains of sleep-deprived and well-rested fruit flies
On top, the brain of a sleep-deprived fly glows orange because of Bruchpilot, a communication protein between brain cells. These bright orange brain areas are associated with learning. On the bottom, a well-rested fly shows lower levels of Bruchpilot, which might make the fly ready to learn after a good night's rest.
Chiara Cirelli, University of Wisconsin-Madison
View Media
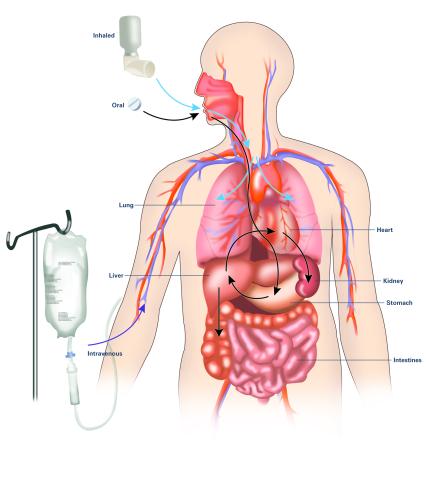
2528: A drug's life in the body (with labels)
2528: A drug's life in the body (with labels)
A drug's life in the body. Medicines taken by mouth (oral) pass through the liver before they are absorbed into the bloodstream. Other forms of drug administration bypass the liver, entering the blood directly. See 2527 for an unlabeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
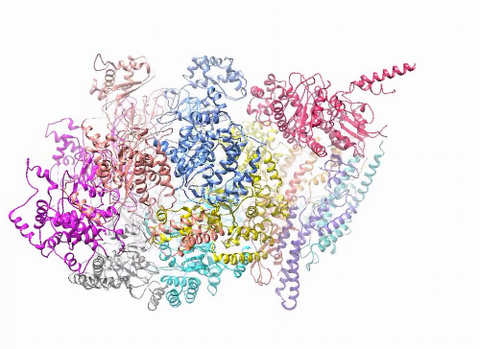
3750: A dynamic model of the DNA helicase protein complex
3750: A dynamic model of the DNA helicase protein complex
This short video shows a model of the DNA helicase in yeast. This DNA helicase has 11 proteins that work together to unwind DNA during the process of copying it, called DNA replication. Scientists used a technique called cryo-electron microscopy (cryo-EM), which allowed them to study the helicase structure in solution rather than in static crystals. Cryo-EM in combination with computer modeling therefore allows researchers to see movements and other dynamic changes in the protein. The cryo-EM approach revealed the helicase structure at much greater resolution than could be obtained before. The researchers think that a repeated motion within the protein as shown in the video helps it move along the DNA strand. To read more about DNA helicase and this proposed mechanism, see this news release by Brookhaven National Laboratory.
Huilin Li, Stony Brook University
View Media
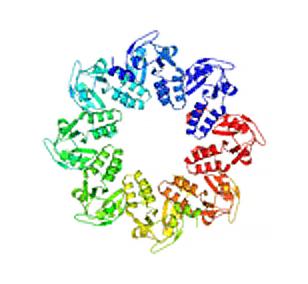
2377: Protein involved in cell division from Mycoplasma pneumoniae
2377: Protein involved in cell division from Mycoplasma pneumoniae
Model of a protein involved in cell division from Mycoplasma pneumoniae. This model, based on X-ray crystallography, revealed a structural domain not seen before. The protein is thought to be involved in cell division and cell wall biosynthesis.
Berkeley Structural Genomics Center, PSI
View Media
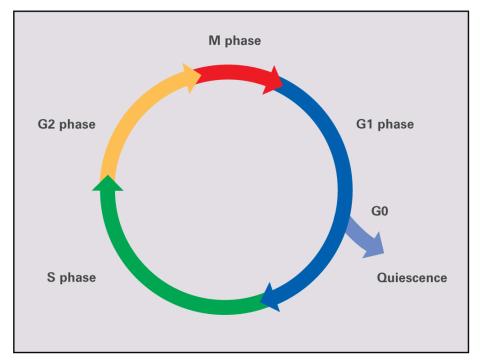
2499: Cell cycle (with labels)
2499: Cell cycle (with labels)
Cells progress through a cycle that consists of phases for growth (G1, S, and G2) and division (M). Cells become quiescent when they exit this cycle (G0). See image 2498 for an unlabeled version of this illustration.
Crabtree + Company
View Media
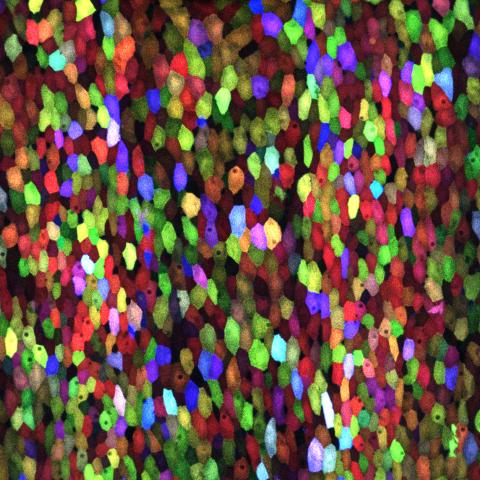
3782: A multicolored fish scale 1
3782: A multicolored fish scale 1
Each of the colored specs in this image is a cell on the surface of a fish scale. To better understand how wounds heal, scientists have inserted genes that make cells brightly glow in different colors into the skin cells of zebrafish, a fish often used in laboratory research. The colors enable the researchers to track each individual cell, for example, as it moves to the location of a cut or scrape over the course of several days. These technicolor fish endowed with glowing skin cells dubbed "skinbow" provide important insight into how tissues recover and regenerate after an injury.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3783.
For more information on skinbow fish, see the Biomedical Beat blog post Visualizing Skin Regeneration in Real Time and a press release from Duke University highlighting this research. Related to image 3783.
Chen-Hui Chen and Kenneth Poss, Duke University
View Media
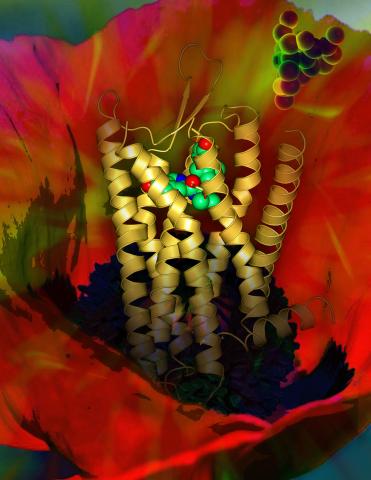
3314: Human opioid receptor structure superimposed on poppy
3314: Human opioid receptor structure superimposed on poppy
Opioid receptors on the surfaces of brain cells are involved in pleasure, pain, addiction, depression, psychosis, and other conditions. The receptors bind to both innate opioids and drugs ranging from hospital anesthetics to opium. Researchers at The Scripps Research Institute, supported by the NIGMS Protein Structure Initiative, determined the first three-dimensional structure of a human opioid receptor, a kappa-opioid receptor. In this illustration, the submicroscopic receptor structure is shown while bound to an agonist (or activator). The structure is superimposed on a poppy flower, the source of opium.
Raymond Stevens, The Scripps Research Institute
View Media
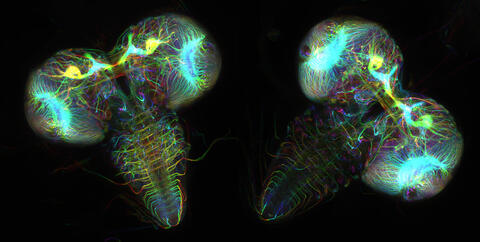
6808: Fruit fly larvae brains showing tubulin
6808: Fruit fly larvae brains showing tubulin
Two fruit fly (Drosophila melanogaster) larvae brains with neurons expressing fluorescently tagged tubulin protein. Tubulin makes up strong, hollow fibers called microtubules that play important roles in neuron growth and migration during brain development. This image was captured using confocal microscopy, and the color indicates the position of the neurons within the brain.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
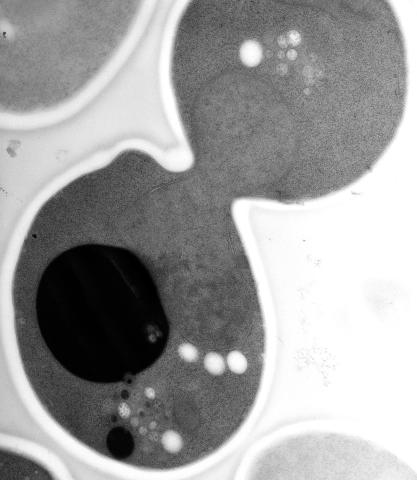
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media
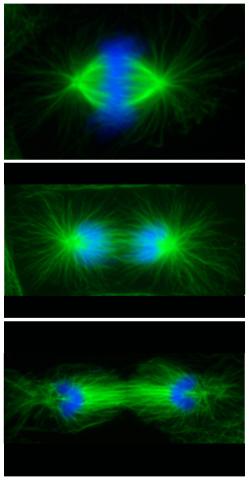
3442: Cell division phases in Xenopus frog cells
3442: Cell division phases in Xenopus frog cells
These images show three stages of cell division in Xenopus XL177 cells, which are derived from tadpole epithelial cells. They are (from top): metaphase, anaphase and telophase. The microtubules are green and the chromosomes are blue. Related to 3443.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison
View Media
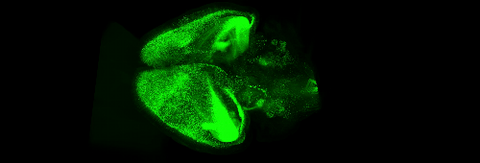
6931: Mouse brain 3
6931: Mouse brain 3
Various views of a mouse brain that was genetically modified so that subpopulations of its neurons glow. Researchers often study mice because they share many genes with people and can shed light on biological processes, development, and diseases in humans.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
This video was captured using a light sheet microscope.
Related to images 6929 and 6930.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media

6756: Honeybees marked with paint
6756: Honeybees marked with paint
Researchers doing behavioral experiments with honeybees sometimes use paint or enamel to give individual bees distinguishing marks. The elaborate social structure and impressive learning and navigation abilities of bees make them good models for behavioral and neurobiological research. Since the sequencing of the honeybee genome, published in 2006, bees have been used increasingly for research into the molecular basis for social interaction and other complex behaviors.
Gene Robinson, University of Illinois at Urbana-Champaign.
View Media
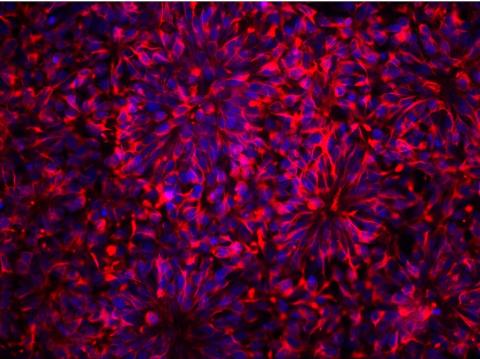
3284: Neurons from human ES cells
3284: Neurons from human ES cells
These neural precursor cells were derived from human embryonic stem cells. The neural cell bodies are stained red, and the nuclei are blue. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Xianmin Zeng lab, Buck Institute for Age Research, via CIRM
View Media

5895: Bioluminescence in a Tube
5895: Bioluminescence in a Tube
Details about the basic biology and chemistry of the ingredients that produce bioluminescence are allowing scientists to harness it as an imaging tool. Credit: Nathan Shaner, Scintillon Institute.
From Biomedical Beat article July 2017: Chasing Fireflies—and Better Cellular Imaging Techniques
From Biomedical Beat article July 2017: Chasing Fireflies—and Better Cellular Imaging Techniques
Nathan Shaner, Scintillon Institute
View Media
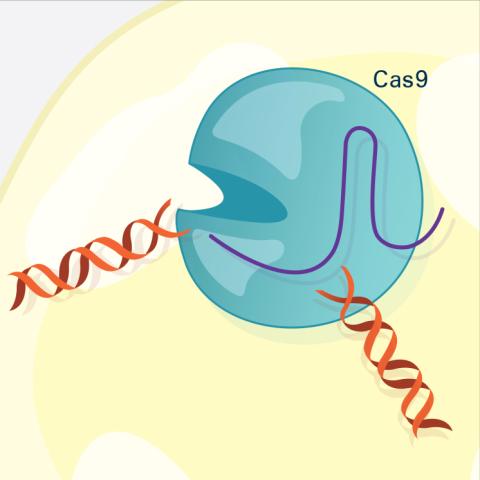
6487: CRISPR Illustration Frame 3
6487: CRISPR Illustration Frame 3
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool. The CRISPR system has two components joined together: a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence) and a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA). In this frame (3 of 4), the Cas9 enzyme cuts both strands of the DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video, and find the full CRIPSR illustration here.
National Institute of General Medical Sciences.
View Media
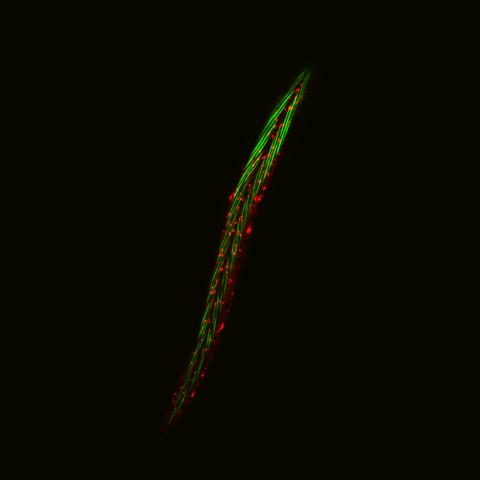
6581: Fluorescent C. elegans showing muscle and ribosomal protein
6581: Fluorescent C. elegans showing muscle and ribosomal protein
C. elegans, a tiny roundworm, with a ribosomal protein glowing red and muscle fibers glowing green. Researchers used these worms to study a molecular pathway that affects aging. The ribosomal protein is involved in protein translation and may play a role in dietary restriction-induced longevity. Image created using confocal microscopy.
View group of roundworms here 6582.
View closeup of roundworms here 6583.
View group of roundworms here 6582.
View closeup of roundworms here 6583.
Jarod Rollins, Mount Desert Island Biological Laboratory.
View Media
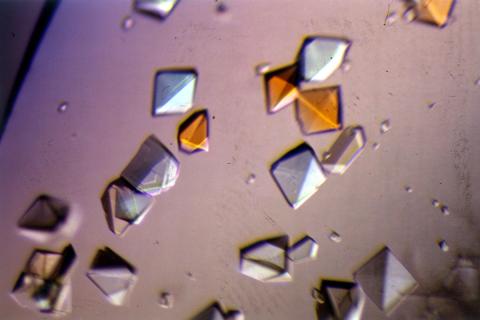
2412: Pig alpha amylase
2412: Pig alpha amylase
Crystals of porcine alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media