Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
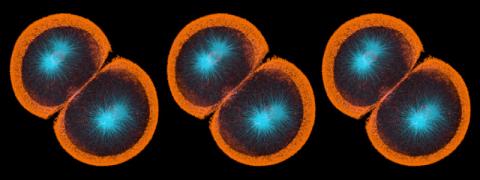
3270: Dopaminergic neurons from ES cells
3270: Dopaminergic neurons from ES cells
Human embryonic stem cells differentiated into dopaminergic neurons, the type that degenerate in Parkinson's disease. Image courtesy of the California Institute for Regenerative Medicine. Related to images 3271 and 3285.
Jeannie Liu, Lab of Jan Nolta, University of California, Davis, via CIRM
View Media
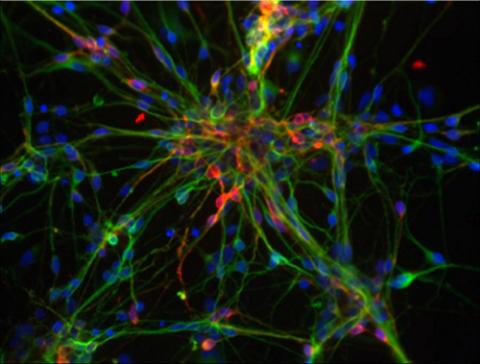
3546: Insulin and protein interact in pancreatic beta cells
3546: Insulin and protein interact in pancreatic beta cells
A large number of proteins interact with the hormone insulin as it is produced in and secreted from the beta cells of the pancreas. In this image, the interactions of TMEM24 protein (green) and insulin (red) in pancreatic beta cells are shown in yellow. More information about the research behind this image can be found in a Biomedical Beat Blog posting from November 2013.
William E. Balch, The Scripps Research Institute
View Media
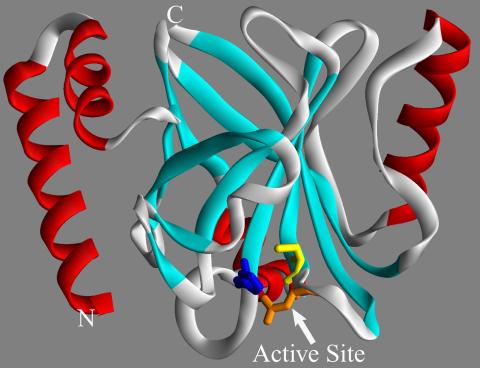
2386: Sortase b from B. anthracis
2386: Sortase b from B. anthracis
Structure of sortase b from the bacterium B. anthracis, which causes anthrax. Sortase b is an enzyme used to rob red blood cells of iron, which the bacteria need to survive.
Midwest Center for Structural Genomics, PSI
View Media
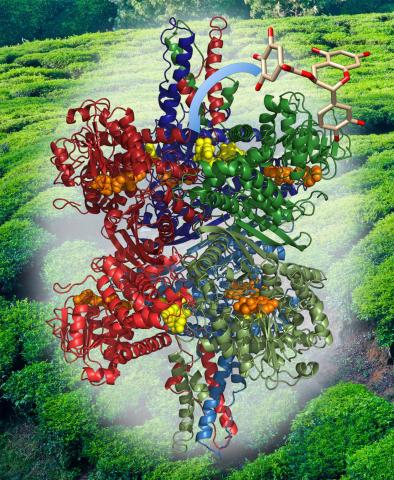
3421: Structure of Glutamate Dehydrogenase
3421: Structure of Glutamate Dehydrogenase
Some children are born with a mutation in a regulatory site on this enzyme that causes them to over-secrete insulin when they consume protein. We found that a compound from green tea (shown in the stick figure and by the yellow spheres on the enzyme) is able to block this hyperactivity when given to animals with this disorder.
Judy Coyle, Donald Danforth Plant Science Center
View Media
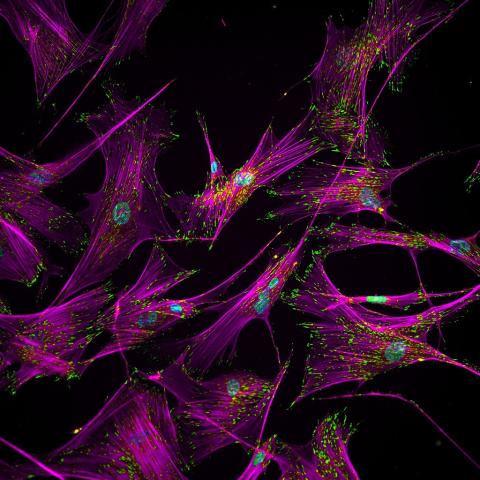
3457: Sticky stem cells
3457: Sticky stem cells
Like a group of barnacles hanging onto a rock, these human cells hang onto a matrix coated glass slide. Actin stress fibers, stained magenta, and the protein vinculin, stained green, make this adhesion possible. The fibroblast nuclei are stained blue.
Ankur Singh and Andrés García, Georgia Institute of Technology
View Media
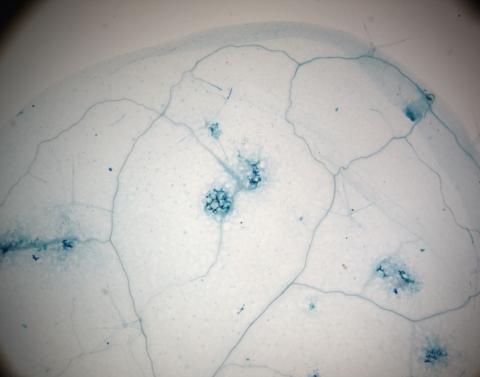
2781: Disease-resistant Arabidopsis leaf
2781: Disease-resistant Arabidopsis leaf
This is a magnified view of an Arabidopsis thaliana leaf a few days after being exposed to the pathogen Hyaloperonospora arabidopsidis. The plant from which this leaf was taken is genetically resistant to the pathogen. The spots in blue show areas of localized cell death where infection occurred, but it did not spread. Compare this response to that shown in Image 2782. Jeff Dangl has been funded by NIGMS to study the interactions between pathogens and hosts that allow or suppress infection.
Jeff Dangl, University of North Carolina, Chapel Hill
View Media
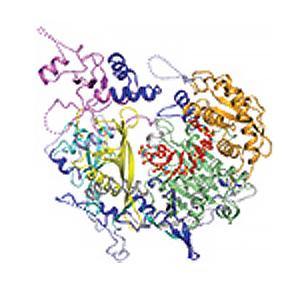
3408: Kluyveromyces polysporus Argonaute bound to guide RNA
3408: Kluyveromyces polysporus Argonaute bound to guide RNA
A segment of siRNA, shown in red, guides a "slicer" protein called Argonaute (multi-colored twists and corkscrews) to the target RNA molecules.
Kotaro Nakanishi and David Weinberg, Massachusetts Institute of Technology
View Media
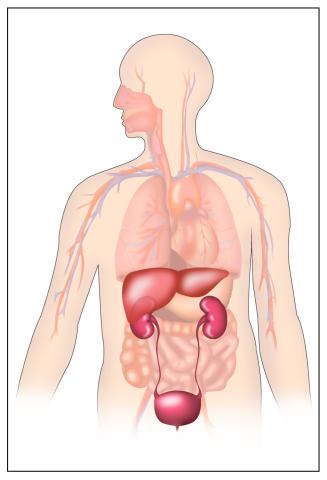
2496: Body toxins
2496: Body toxins
Body organs such as the liver and kidneys process chemicals and toxins. These "target" organs are susceptible to damage caused by these substances. See image 2497 for a labeled version of this illustration.
Crabtree + Company
View Media
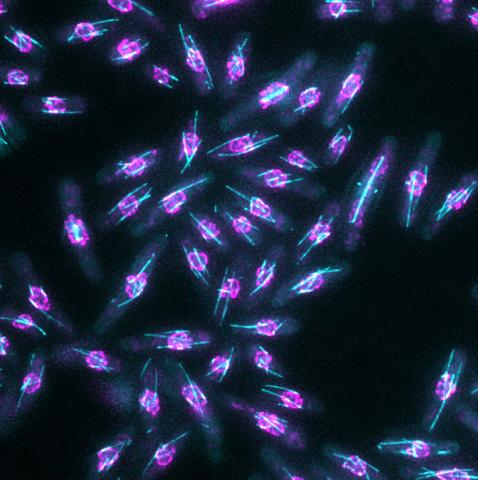
6798: Yeast cells with nuclear envelopes and tubulin
6798: Yeast cells with nuclear envelopes and tubulin
Yeast cells with nuclear envelopes shown in magenta and tubulin shown in light blue. The nuclear envelope defines the borders of the nucleus, which houses DNA. Tubulin is a protein that makes up microtubules—strong, hollow fibers that provide structure to cells and help direct chromosomes during cell division. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6797, and videos 6795 and 6796.
Related to images 6791, 6792, 6793, 6794, 6797, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
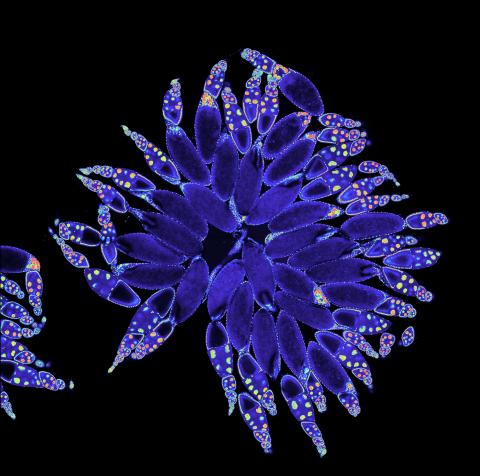
3607: Fruit fly ovary
3607: Fruit fly ovary
A fruit fly ovary, shown here, contains as many as 20 eggs. Fruit flies are not merely tiny insects that buzz around overripe fruit—they are a venerable scientific tool. Research on the flies has shed light on many aspects of human biology, including biological rhythms, learning, memory, and neurodegenerative diseases. Another reason fruit flies are so useful in a lab (and so successful in fruit bowls) is that they reproduce rapidly. About three generations can be studied in a single month.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Related to image 3656. This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Denise Montell, Johns Hopkins University and University of California, Santa Barbara
View Media
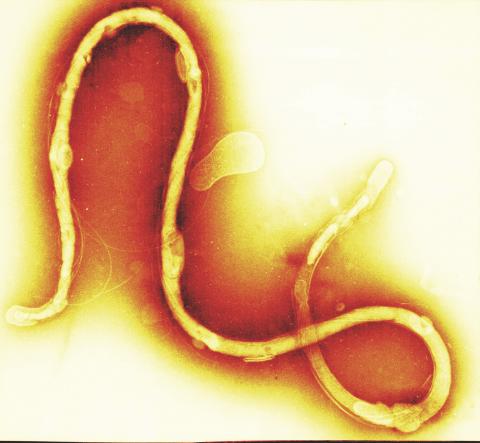
1241: Borrelia burgdorferi
1241: Borrelia burgdorferi
Borrelia burgdorferi is a spirochete, a class of long, slender bacteria that typically take on a coiled shape. Infection with this bacterium causes Lyme disease.
Tina Weatherby Carvalho, University of Hawaii at Manoa
View Media

6752: Petri dish
6752: Petri dish
The white circle in this image is a Petri dish, named for its inventor, Julius Richard Petri. These dishes are one of the most common pieces of equipment in biology labs, where researchers use them to grow cells.
H. Robert Horvitz and Dipon Ghosh, Massachusetts Institute of Technology.
View Media
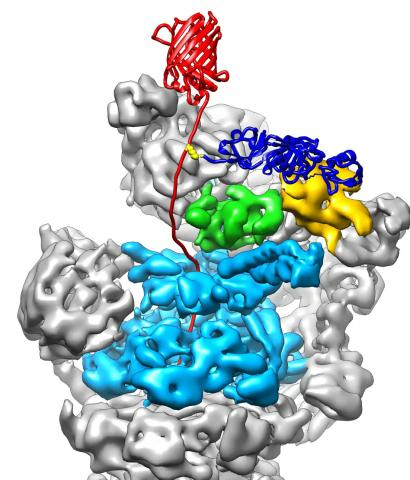
3763: The 26S proteasome engages with a protein substrate
3763: The 26S proteasome engages with a protein substrate
The proteasome is a critical multiprotein complex in the cell that breaks down and recycles proteins that have become damaged or are no longer needed. This illustration shows a protein substrate (red) that is bound through its ubiquitin chain (blue) to one of the ubiquitin receptors of the proteasome (Rpn10, yellow). The substrate's flexible engagement region gets engaged by the AAA+ motor of the proteasome (cyan), which initiates mechanical pulling, unfolding and movement of the protein into the proteasome's interior for cleavage into small shorter protein pieces called peptides. During movement of the substrate, its ubiquitin modification gets cleaved off by the deubiquitinase Rpn11 (green), which sits directly above the entrance to the AAA+ motor pore and acts as a gatekeeper to ensure efficient ubiquitin removal, a prerequisite for fast protein breakdown by the 26S proteasome. Related to video 3764.
Andreas Martin, HHMI
View Media
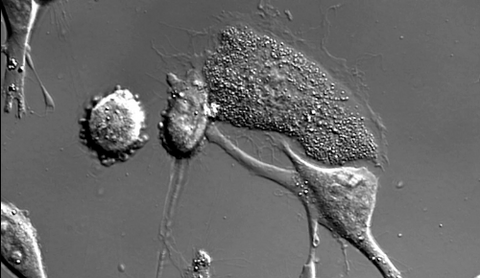
6966: Dying melanoma cells
6966: Dying melanoma cells
Melanoma (skin cancer) cells undergoing programmed cell death, also called apoptosis. This process was triggered by raising the pH of the medium that the cells were growing in. Melanoma in people cannot be treated by raising pH because that would also kill healthy cells. This video was taken using a differential interference contrast (DIC) microscope.
Dylan T. Burnette, Vanderbilt University School of Medicine.
View Media

7012: Adult Hawaiian bobtail squid burying in the sand
7012: Adult Hawaiian bobtail squid burying in the sand
Each morning, the nocturnal Hawaiian bobtail squid, Euprymna scolopes, hides from predators by digging into the sand. At dusk, it leaves the sand again to hunt.
Related to image 7010 and 7011.
Related to image 7010 and 7011.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
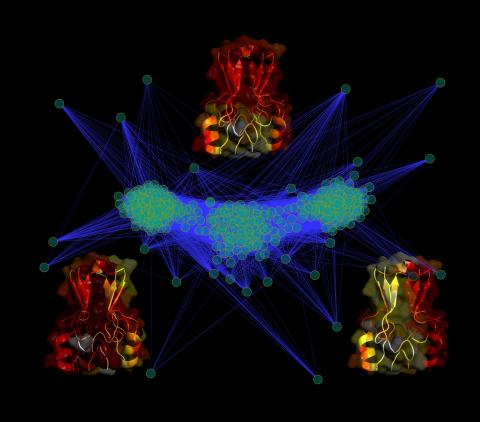
3295: Cluster analysis of mysterious protein
3295: Cluster analysis of mysterious protein
Researchers use cluster analysis to study protein shape and function. Each green circle represents one potential shape of the protein mitoNEET. The longer the blue line between two circles, the greater the differences between the shapes. Most shapes are similar; they fall into three clusters that are represented by the three images of the protein. From a Rice University news release. Graduate student Elizabeth Baxter and Patricia Jennings, professor of chemistry and biochemistry at UCSD, collaborated with José Onuchic, a physicist at Rice University, on this work.
Patricia Jennings and Elizabeth Baxter, University of California, San Diego
View Media
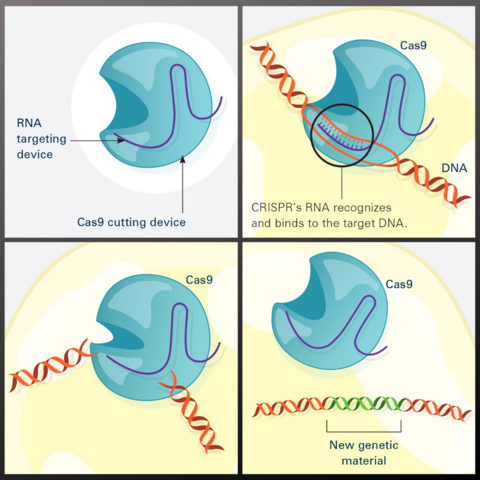
7036: CRISPR Illustration
7036: CRISPR Illustration
This illustration shows, in simplified terms, how the CRISPR-Cas9 system can be used as a gene-editing tool.
Frame 1 shows the two components of the CRISPR system: a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA), and a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence).
In frame 2, the CRISPR machine locates the target DNA sequence once inserted into a cell.
In frame 3, the Cas9 enzyme cuts both strands of the DNA.
Frame 4 shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video.
Download the individual frames: Frame 1, Frame 2, Frame 3, and Frame 4.
Frame 1 shows the two components of the CRISPR system: a strong cutting device (an enzyme called Cas9 that can cut through a double strand of DNA), and a finely tuned targeting device (a small strand of RNA programmed to look for a specific DNA sequence).
In frame 2, the CRISPR machine locates the target DNA sequence once inserted into a cell.
In frame 3, the Cas9 enzyme cuts both strands of the DNA.
Frame 4 shows a repaired DNA strand with new genetic material that researchers can introduce, which the cell automatically incorporates into the gap when it repairs the broken DNA.
For an explanation and overview of the CRISPR-Cas9 system, see the iBiology video.
Download the individual frames: Frame 1, Frame 2, Frame 3, and Frame 4.
National Institute of General Medical Sciences.
View Media
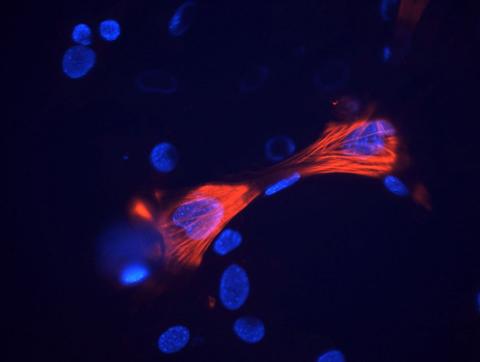
3289: Smooth muscle from mouse stem cells
3289: Smooth muscle from mouse stem cells
These smooth muscle cells were derived from mouse neural crest stem cells. Red indicates smooth muscle proteins, blue indicates nuclei. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Deepak Srivastava, Gladstone Institutes, via CIRM
View Media

2715: Glow-in-the-dark salamanders
2715: Glow-in-the-dark salamanders
These six-month-old axolotls, a kind of salamander, glow green and blue under ultraviolet light. That's because they were genetically modified to make harmless green fluorescent protein, or GFP. Like X-ray vision, GFP lets you see inside the axolotls as they hang out in their aquarium. GFP not only can reveal internal structures in living organisms, but it also can light up specific cells and even proteins within a cell. That allows scientists to identify and track things like cancer cells.
View Media

2714: Stretch detectors
2714: Stretch detectors
Muscles stretch and contract when we walk, and skin splits open and knits back together when we get a paper cut. To study these contractile forces, researchers built a three-dimensional scaffold that mimics tissue in an organism. Researchers poured a mixture of cells and elastic collagen over microscopic posts in a dish. Then they studied how the cells pulled and released the posts as they formed a web of tissue. To measure forces between posts, the researchers developed a computer model. Their findings--which show that contractile forces vary throughout the tissue--could have a wide range of medical applications.
Christopher Chen, University of Pennsylvania
View Media
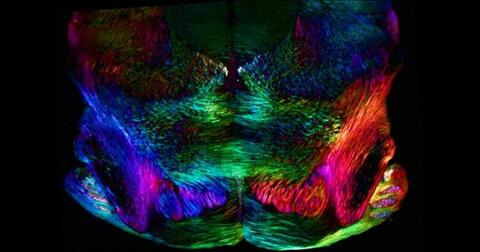
6901: Mouse brain slice showing nerve cells
6901: Mouse brain slice showing nerve cells
A 20-µm thick section of mouse midbrain. The nerve cells are transparent and weren’t stained. Instead, the color is generated by interaction of white polarized light with the molecules in the cells and indicates their orientation.
The image was obtained with a polychromatic polarizing microscope that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about the microscopy that produced this image can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
The image was obtained with a polychromatic polarizing microscope that shows the polychromatic birefringent image with hue corresponding to the slow axis orientation. More information about the microscopy that produced this image can be found in the Scientific Reports paper “Polychromatic Polarization Microscope: Bringing Colors to a Colorless World” by Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
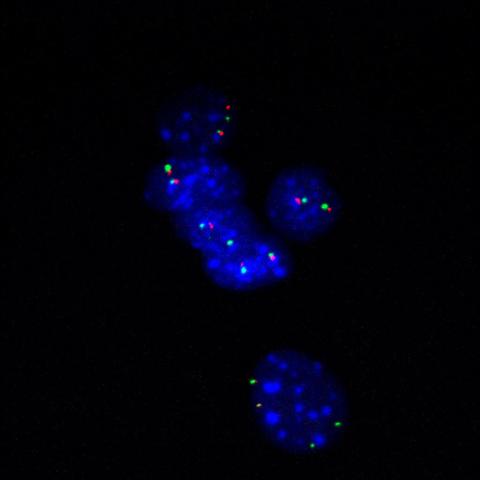
3296: Fluorescence in situ hybridization (FISH) in mouse ES cells shows DNA interactions
3296: Fluorescence in situ hybridization (FISH) in mouse ES cells shows DNA interactions
Researchers used fluorescence in situ hybridization (FISH) to confirm the presence of long range DNA-DNA interactions in mouse embryonic stem cells. Here, two loci labeled in green (Oct4) and red that are 13 Mb apart on linear DNA are frequently found to be in close proximity. DNA-DNA colocalizations like this are thought to both reflect and contribute to cell type specific gene expression programs.
Kathrin Plath, University of California, Los Angeles
View Media
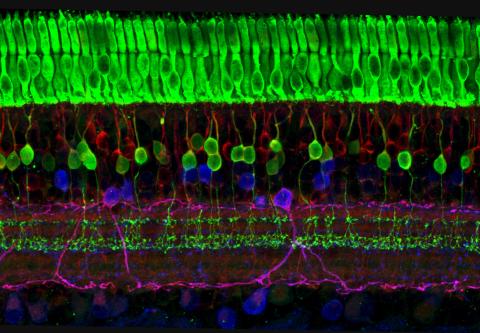
3635: The eye uses many layers of nerve cells to convert light into sight
3635: The eye uses many layers of nerve cells to convert light into sight
This image captures the many layers of nerve cells in the retina. The top layer (green) is made up of cells called photoreceptors that convert light into electrical signals to relay to the brain. The two best-known types of photoreceptor cells are rod- and cone-shaped. Rods help us see under low-light conditions but can't help us distinguish colors. Cones don't function well in the dark but allow us to see vibrant colors in daylight.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Wei Li, National Eye Institute, National Institutes of Health
View Media
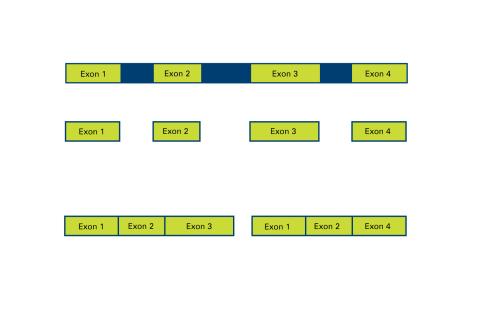
2552: Alternative splicing
2552: Alternative splicing
Arranging exons in different patterns, called alternative splicing, enables cells to make different proteins from a single gene. See image 2553 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
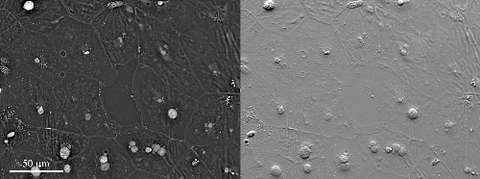
6899: Epithelial cell migration
6899: Epithelial cell migration
High-resolution time lapse of epithelial (skin) cell migration and wound healing. It shows an image taken every 13 seconds over the course of almost 14 minutes. The images were captured with quantitative orientation-independent differential interference contrast (DIC) microscope (left) and a conventional DIC microscope (right).
More information about the research that produced this video can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
More information about the research that produced this video can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
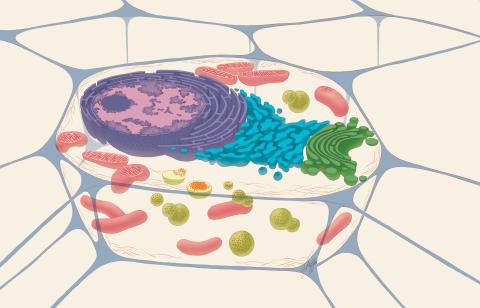
1274: Animal cell
1274: Animal cell
A typical animal cell, sliced open to reveal a cross-section of organelles.
Judith Stoffer
View Media
6776: Tracking cells in a gastrulating zebrafish embryo
6776: Tracking cells in a gastrulating zebrafish embryo
During development, a zebrafish embryo is transformed from a ball of cells into a recognizable body plan by sweeping convergence and extension cell movements. This process is called gastrulation. Each line in this video represents the movement of a single zebrafish embryo cell over the course of 3 hours. The video was created using time-lapse confocal microscopy. Related to image 6775.
Liliana Solnica-Krezel, Washington University School of Medicine in St. Louis.
View Media

2747: Cell division with late aligning chromosomes
2747: Cell division with late aligning chromosomes
This video shows an instance of abnormal mitosis where chromosomes are late to align. The video demonstrates the spindle checkpoint in action: just one unaligned chromosome can delay anaphase and the completion of mitosis. The cells shown are S3 tissue cultured cells from Xenopus laevis, African clawed frog.
Gary Gorbsky, Oklahoma Medical Research Foundation
View Media
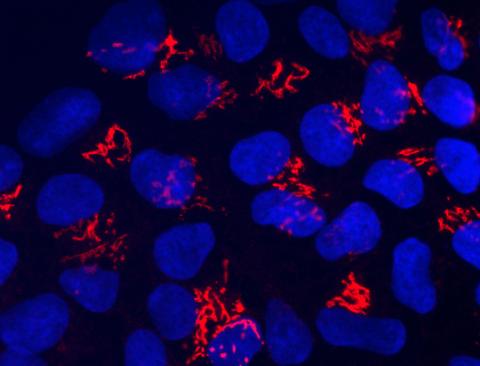
3341: Suicidal Stem Cells
3341: Suicidal Stem Cells
Embryonic stem cells store pre-activated Bax (red) in the Golgi, near the nucleus (blue). Featured in the June 21, 2012, issue of Biomedical Beat.
Mohanish Deshmukh
View Media

3567: RSV-Infected Cell
3567: RSV-Infected Cell
Viral RNA (red) in an RSV-infected cell. More information about the research behind this image can be found in a Biomedical Beat Blog posting from January 2014.
Eric Alonas and Philip Santangelo, Georgia Institute of Technology and Emory University
View Media
2474: Dinosaur evolutionary tree
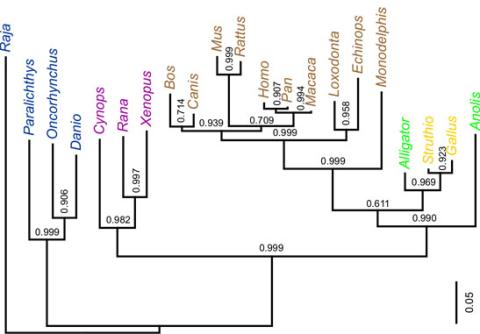
2474: Dinosaur evolutionary tree
Analysis of 68 million-year-old collagen molecule fragments preserved in a T. rex femur confirmed what paleontologists have said for decades: Dinosaurs are close relatives of chickens, ostriches, and to a lesser extent, alligators. A Harvard University research team, including NIGMS-supported postdoctoral research fellow Chris Organ, used sophisticated statistical and computational tools to compare the ancient protein to ones from 21 living species. Because evolutionary processes produce similarities across species, the methods and results may help illuminate other areas of the evolutionary tree. Featured in the May 21, 2008 Biomedical Beat.
Chris Organ, Harvard University
View Media
3400: Small blood vessels in a mouse retina
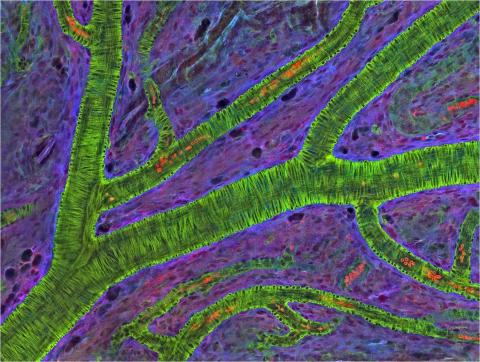
3400: Small blood vessels in a mouse retina
Blood vessels at the back of the eye (retina) are used to diagnose glaucoma and diabetic eye disease. They also display characteristic changes in people with high blood pressure. In the image, the vessels appear green. It's not actually the vessels that are stained green, but rather filaments of a protein called actin that wraps around the vessels. Most of the red blood cells were replaced by fluid as the tissue was prepared for the microscope. The tiny red dots are red blood cells that remain in the vessels. The image was captured using confocal and 2-photon excitation microscopy for a project related to neurofibromatosis.
National Center for Microscopy and Imaging Research
View Media
2327: Neural development
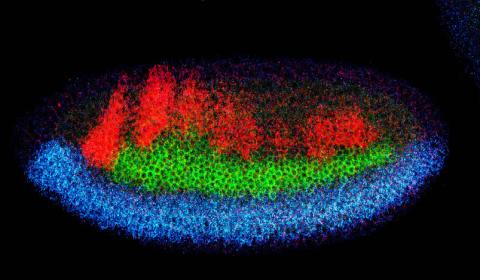
2327: Neural development
Using techniques that took 4 years to design, a team of developmental biologists showed that certain proteins can direct the subdivision of fruit fly and chicken nervous system tissue into the regions depicted here in blue, green, and red. Molecules called bone morphogenetic proteins (BMPs) helped form this fruit fly embryo. While scientists knew that BMPs play a major role earlier in embryonic development, they didn't know how the proteins help organize nervous tissue. The findings suggest that BMPs are part of an evolutionarily conserved mechanism for organizing the nervous system. The National Institute of Neurological Disorders and Stroke also supported this work.
Mieko Mizutani and Ethan Bier, University of California, San Diego, and Henk Roelink, University of Washington
View Media
1018: Lily mitosis 12
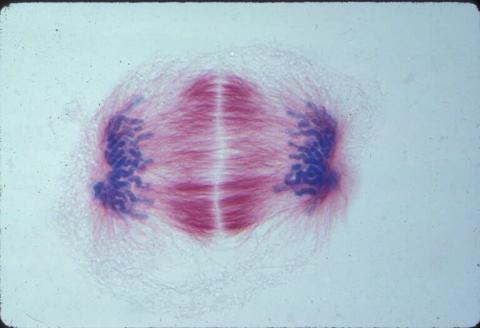
1018: Lily mitosis 12
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible near the end of a round of mitosis.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Related to images 1010, 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media
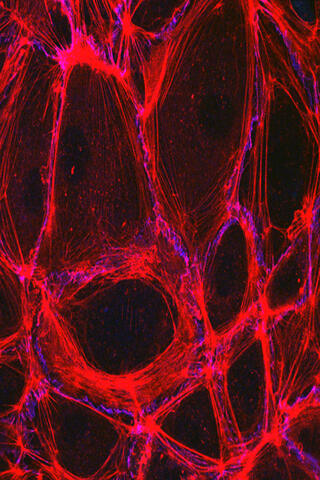
3633: Cells lining the blood vessel walls
3633: Cells lining the blood vessel walls
The structure of the endothelium, the thin layer of cells that line our arteries and veins, is visible here. The endothelium is like a gatekeeper, controlling the movement of materials into and out of the bloodstream. Endothelial cells are held tightly together by specialized proteins that function like strong ropes (red) and others that act like cement (blue).
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Christopher V. Carman and Roberta Martinelli, Harvard Medical School.
View Media
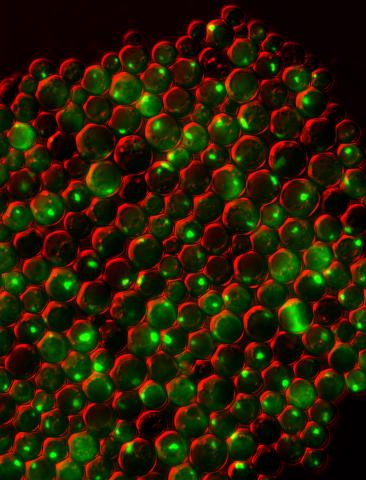
3550: Protein clumping in zinc-deficient yeast cells
3550: Protein clumping in zinc-deficient yeast cells
The green spots in this image are clumps of protein inside yeast cells that are deficient in both zinc and a protein called Tsa1 that prevents clumping. Protein clumping plays a role in many diseases, including Parkinson's and Alzheimer's, where proteins clump together in the brain. Zinc deficiency within a cell can cause proteins to mis-fold and eventually clump together. Normally, in yeast, Tsa1 codes for so-called "chaperone proteins" which help proteins in stressed cells, such as those with a zinc deficiency, fold correctly. The research behind this image was published in 2013 in the Journal of Biological Chemistry.
Colin MacDiarmid and David Eide, University of Wisconsin--Madison
View Media

3733: A molecular interaction network in yeast 3
3733: A molecular interaction network in yeast 3
The image visualizes a part of the yeast molecular interaction network. The lines in the network represent connections among genes (shown as little dots) and different-colored networks indicate subnetworks, for instance, those in specific locations or pathways in the cell. Researchers use gene or protein expression data to build these networks; the network shown here was visualized with a program called Cytoscape. By following changes in the architectures of these networks in response to altered environmental conditions, scientists can home in on those genes that become central "hubs" (highly connected genes), for example, when a cell encounters stress. They can then further investigate the precise role of these genes to uncover how a cell's molecular machinery deals with stress or other factors. Related to images 3730 and 3732.
Keiichiro Ono, UCSD
View Media
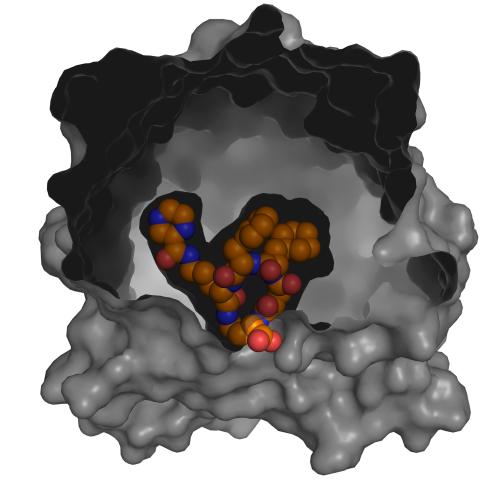
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
3417: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 5
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to images 3413, 3414, 3415, 3416, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
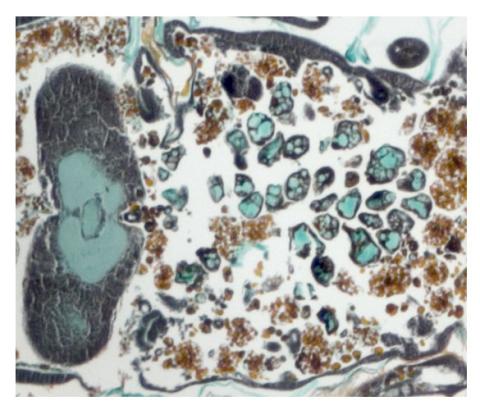
2759: Cross section of a Drosophila melanogaster pupa lacking Draper
2759: Cross section of a Drosophila melanogaster pupa lacking Draper
In the absence of the engulfment receptor Draper, salivary gland cells (light blue) persist in the thorax of a developing Drosophila melanogaster pupa. See image 2758 for a cross section of a normal pupa that does express Draper.
Christina McPhee and Eric Baehrecke, University of Massachusetts Medical School
View Media
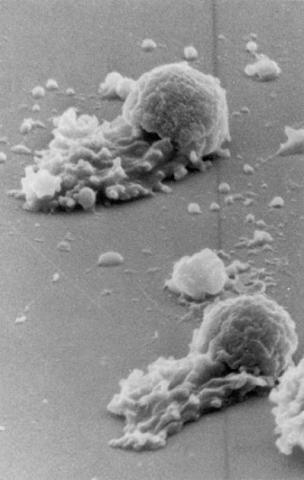
3489: Worm sperm
3489: Worm sperm
To develop a system for studying cell motility in unnatrual conditions -- a microscope slide instead of the body -- Tom Roberts and Katsuya Shimabukuro at Florida State University disassembled and reconstituted the motility parts used by worm sperm cells.
Tom Roberts, Florida State University
View Media
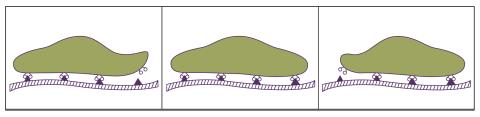
2502: Focal adhesions
2502: Focal adhesions
Cells walk along body surfaces via tiny "feet," called focal adhesions, that connect with the extracellular matrix. See image 2503 for a labeled version of this illustration.
Crabtree + Company
View Media
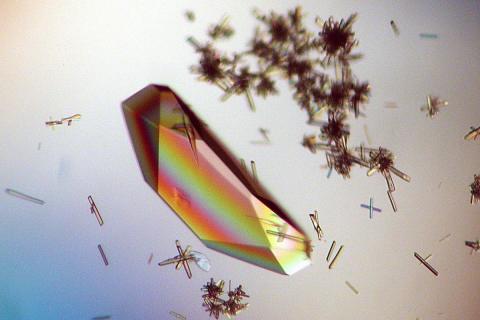
2396: Hen egg lysozyme (1)
2396: Hen egg lysozyme (1)
Crystals of hen egg lysozyme protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
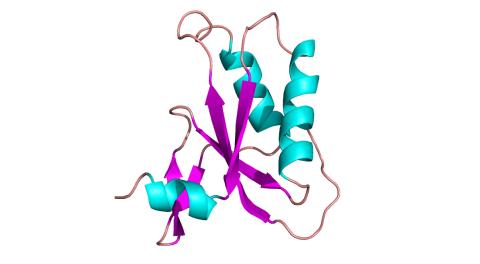
3428: Antitoxin GhoS (Illustration 2)
3428: Antitoxin GhoS (Illustration 2)
Structure of the bacterial antitoxin protein GhoS. GhoS inhibits the production of a bacterial toxin, GhoT, which can contribute to antibiotic resistance. GhoS is the first known bacterial antitoxin that works by cleaving the messenger RNA that carries the instructions for making the toxin. More information can be found in the paper: Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012 Oct;8(10):855-61. Related to 3427.
Rebecca Page and Wolfgang Peti, Brown University and Thomas K. Wood, Pennsylvania State University
View Media
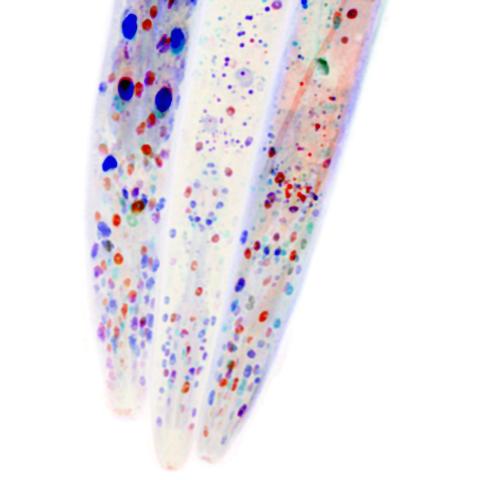
6534: Mosaicism in C. elegans (White Background)
6534: Mosaicism in C. elegans (White Background)
In the worm C. elegans, double-stranded RNA made in neurons can silence matching genes in a variety of cell types through the transport of RNA between cells. The head region of three worms that were genetically modified to express a fluorescent protein were imaged and the images were color-coded based on depth. The worm on the left lacks neuronal double-stranded RNA and thus every cell is fluorescent. In the middle worm, the expression of the fluorescent protein is silenced by neuronal double-stranded RNA and thus most cells are not fluorescent. The worm on the right lacks an enzyme that amplifies RNA for silencing. Surprisingly, the identities of the cells that depend on this enzyme for gene silencing are unpredictable. As a result, worms of identical genotype are nevertheless random mosaics for how the function of gene silencing is carried out. For more, see journal article and press release. Related to image 6532.
Snusha Ravikumar, Ph.D., University of Maryland, College Park, and Antony M. Jose, Ph.D., University of Maryland, College Park
View Media
2430: Fruit fly retina 01
2430: Fruit fly retina 01
Image showing rhabdomeres (red), the light-sensitive structures in the fruit fly retina, and rhodopsin-4 (blue), a light-sensing molecule.
Hermann Steller, Rockefeller University
View Media
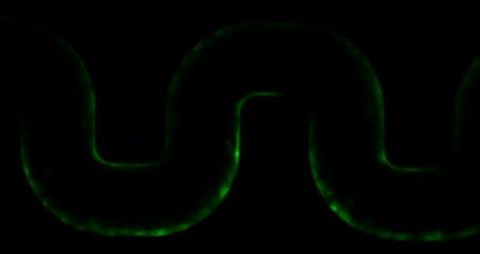
3446: Biofilm blocking fluid flow
3446: Biofilm blocking fluid flow
This time-lapse movie shows that bacterial communities called biofilms can create blockages that prevent fluid flow in devices such as stents and catheters over a period of about 56 hours. This video was featured in a news release from Princeton University.
Bonnie Bassler, Princeton University
View Media
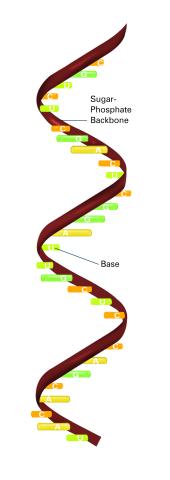
2555: RNA strand (with labels)
2555: RNA strand (with labels)
Ribonucleic acid (RNA) has a sugar-phosphate backbone and the bases adenine (A), cytosine (C), guanine (G), and uracil (U). Featured in The New Genetics.
See image 2554 for an unlabeled version of this illustration.
See image 2554 for an unlabeled version of this illustration.
Crabtree + Company
View Media

2375: Protein purification robot
2375: Protein purification robot
Irina Dementieva, a biochemist, and Youngchang Kim, a biophysicist and crystallographer, work with the first robot of its type in the U.S. to automate protein purification. The robot, which is housed in a refrigerator, is an integral part of the Midwest Structural Genomics Center's plan to automate the protein crystallography process.
Midwest Center for Structural Genomics
View Media
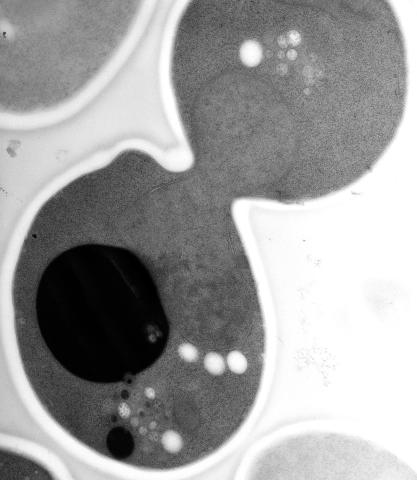
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media