Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
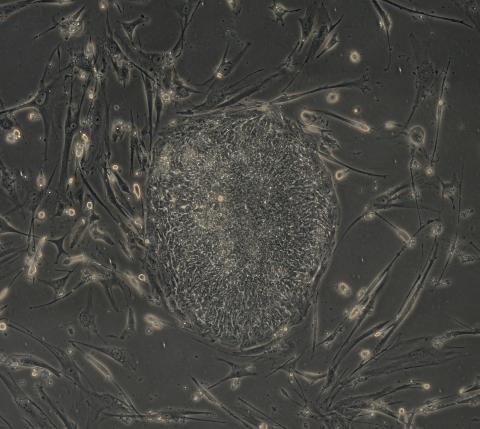
2605: Induced stem cells from adult skin 03
2605: Induced stem cells from adult skin 03
The human skin cells pictured contain genetic modifications that make them pluripotent, essentially equivalent to embryonic stem cells. A scientific team from the University of Wisconsin-Madison including researchers Junying Yu, James Thomson, and their colleagues produced the transformation by introducing a set of four genes into human fibroblasts, skin cells that are easy to obtain and grow in culture.
James Thomson, University of Wisconsin-Madison
View Media

3425: Red Poppy
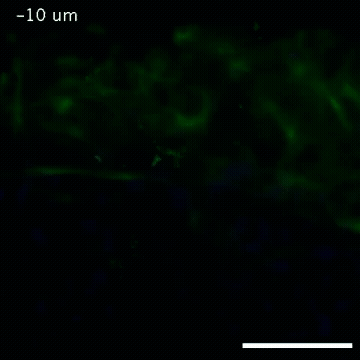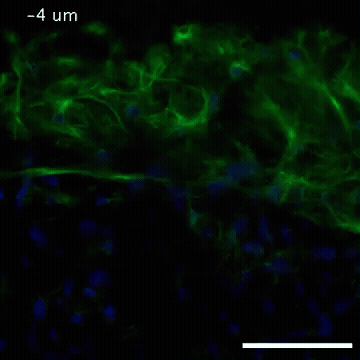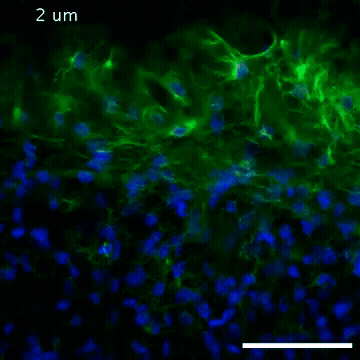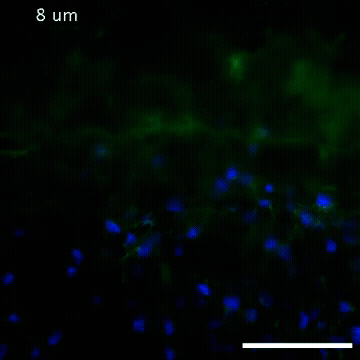
2809: Vimentin in a quail embryo
2809: Vimentin in a quail embryo
Video of high-resolution confocal images depicting vimentin immunofluorescence (green) and nuclei (blue) at the edge of a quail embryo yolk. These images were obtained as part of a study to understand cell migration in embryos. An NIGMS grant to Professor Garcia was used to purchase the confocal microscope that collected these images. Related to images 2807 and 2808.
Andrés Garcia, Georgia Tech
View Media
2767: Research mentor and student
2767: Research mentor and student
A research mentor (Lori Eidson) and student (Nina Waldron, on the microscope) were 2009 members of the BRAIN (Behavioral Research Advancements In Neuroscience) program at Georgia State University in Atlanta. This program is an undergraduate summer research experience funded in part by NIGMS.
Elizabeth Weaver, Georgia State University
View Media
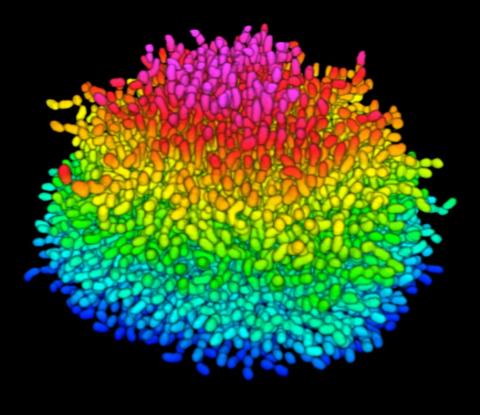
5825: A Growing Bacterial Biofilm
5825: A Growing Bacterial Biofilm
A growing Vibrio cholerae (cholera) biofilm. Cholera bacteria form colonies called biofilms that enable them to resist antibiotic therapy within the body and other challenges to their growth.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Each slightly curved comma shape represents an individual bacterium from assembled confocal microscopy images. Different colors show each bacterium’s position in the biofilm in relation to the surface on which the film is growing.
Jing Yan, Ph.D., and Bonnie Bassler, Ph.D., Department of Molecular Biology, Princeton University, Princeton, NJ.
View Media

2369: Protein purification robot in action 01
2369: Protein purification robot in action 01
A robot is transferring 96 purification columns to a vacuum manifold for subsequent purification procedures.
The Northeast Collaboratory for Structural Genomics
View Media
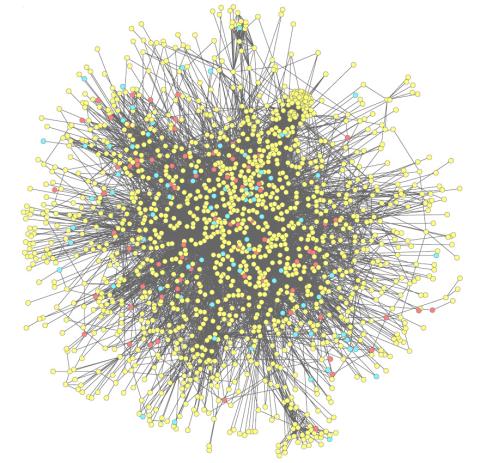
2743: Molecular interactions
2743: Molecular interactions
This network map shows molecular interactions (yellow) associated with a congenital condition that causes heart arrhythmias and the targets for drugs that alter these interactions (red and blue).
Ravi Iyengar, Mount Sinai School of Medicine
View Media
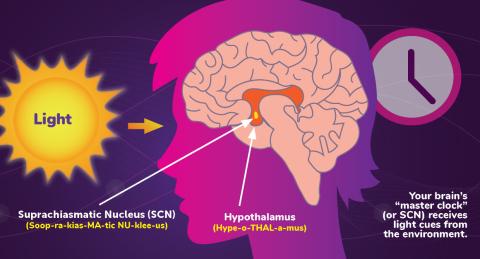
6613: Circadian rhythms and the SCN
6613: Circadian rhythms and the SCN
Circadian rhythms are physical, mental, and behavioral changes that follow a 24-hour cycle. Circadian rhythms are influenced by light and regulated by the brain’s suprachiasmatic nucleus (SCN), sometimes referred to as a master clock. Learn more in NIGMS’ circadian rhythms fact sheet. See 6614 for the Spanish version of this infographic.
NIGMS
View Media
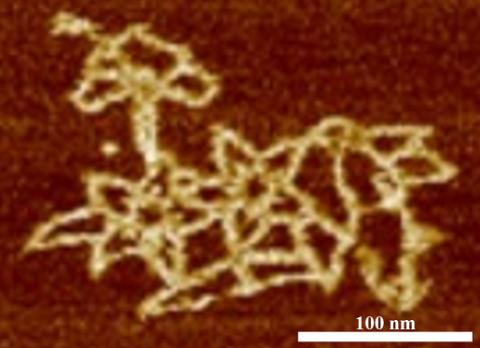
3690: Microscopy image of bird-and-flower DNA origami
3690: Microscopy image of bird-and-flower DNA origami
An atomic force microscopy image shows DNA folded into an intricate, computer-designed structure. Image is featured on Biomedical Beat blog post Cool Image: DNA Origami. See also related image 3689 .
Hao Yan, Arizona State University
View Media
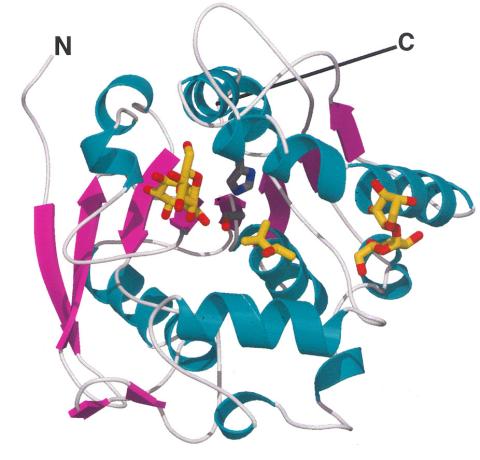
2378: Most abundant protein in M. tuberculosis
2378: Most abundant protein in M. tuberculosis
Model of a protein, antigen 85B, that is the most abundant protein exported by Mycobacterium tuberculosis, which causes most cases of tuberculosis. Antigen 85B is involved in building the bacterial cell wall and is an attractive drug target. Based on its structure, scientists have suggested a new class of antituberculous drugs.
Mycobacterium Tuberculosis Center, PSI
View Media
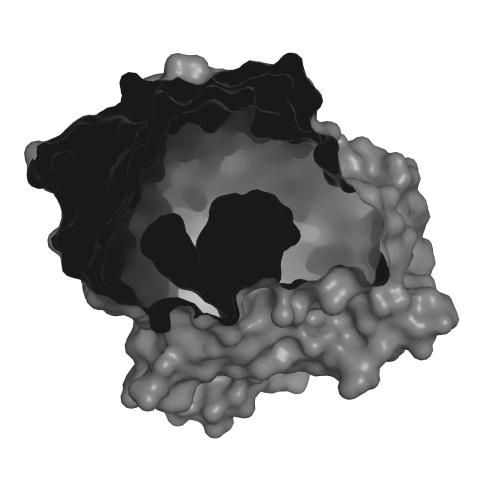
3414: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 2
3414: X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor 2
X-ray co-crystal structure of Src kinase bound to a DNA-templated macrocycle inhibitor. Related to 3413, 3415, 3416, 3417, 3418, and 3419.
Markus A. Seeliger, Stony Brook University Medical School and David R. Liu, Harvard University
View Media
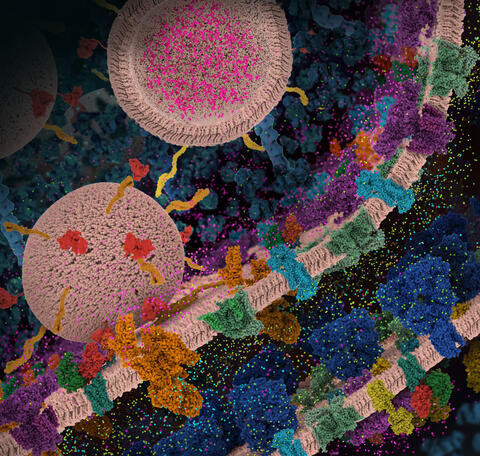
6992: Molecular view of glutamatergic synapse
6992: Molecular view of glutamatergic synapse
This illustration highlights spherical pre-synaptic vesicles that carry the neurotransmitter glutamate. The presynaptic and postsynaptic membranes are shown with proteins relevant for transmitting and modulating the neuronal signal.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
PDB 101’s Opioids and Pain Signaling video explains how glutamatergic synapses are involved in the process of pain signaling.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
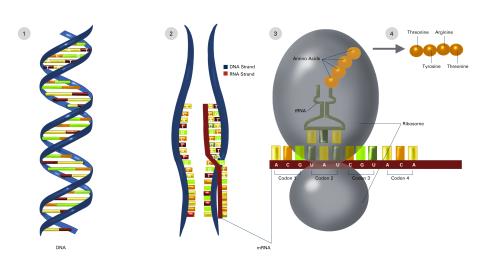
2549: Central dogma, illustrated (with labels and numbers for stages)
2549: Central dogma, illustrated (with labels and numbers for stages)
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a version of this illustration that isn't numbered and 2547 for a an entirely unlabeled version. Featured in The New Genetics.
Crabtree + Company
View Media
1082: Natcher Building 02
1082: Natcher Building 02
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
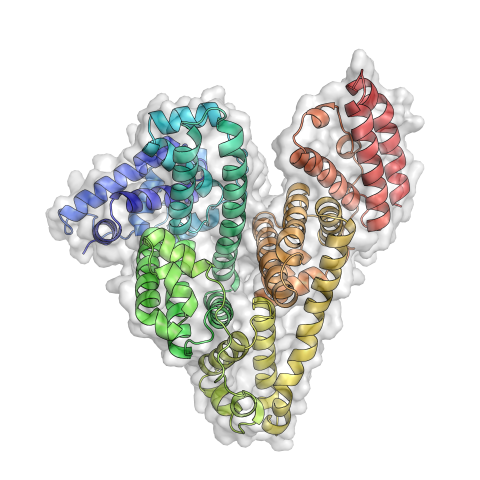
3746: Serum albumin structure 3
3746: Serum albumin structure 3
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3744 and 3745.
Related to entries 3744 and 3745.
Wladek Minor, University of Virginia
View Media
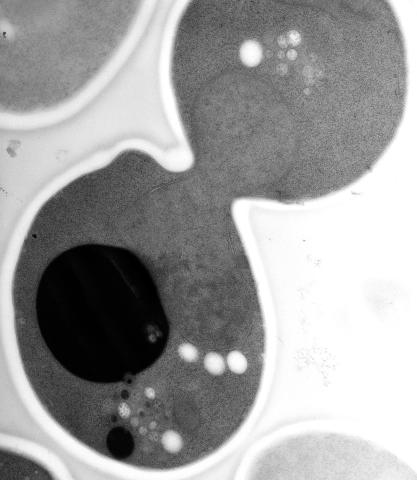
5770: EM of yeast cell division
5770: EM of yeast cell division
Cell division is an incredibly coordinated process. It not only ensures that the new cells formed during this event have a full set of chromosomes, but also that they are endowed with all the cellular materials, including proteins, lipids and small functional compartments called organelles, that are required for normal cell activity. This proper apportioning of essential cell ingredients helps each cell get off to a running start.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
This image shows an electron microscopy (EM) thin section taken at 10,000x magnification of a dividing yeast cell over-expressing the protein ubiquitin, which is involved in protein degradation and recycling. The picture features mother and daughter endosome accumulations (small organelles with internal vesicles), a darkly stained vacuole and a dividing nucleus in close contact with a cadre of lipid droplets (unstained spherical bodies). Other dynamic events are also visible, such as spindle microtubules in the nucleus and endocytic pits at the plasma membrane.
These extensive details were revealed thanks to a preservation method involving high-pressure freezing, freeze-substitution and Lowicryl HM20 embedding.
Matthew West and Greg Odorizzi, University of Colorado
View Media
1056: Skin cross-section
1056: Skin cross-section
Cross-section of skin anatomy shows layers and different tissue types.
National Institutes of Health Medical Arts
View Media
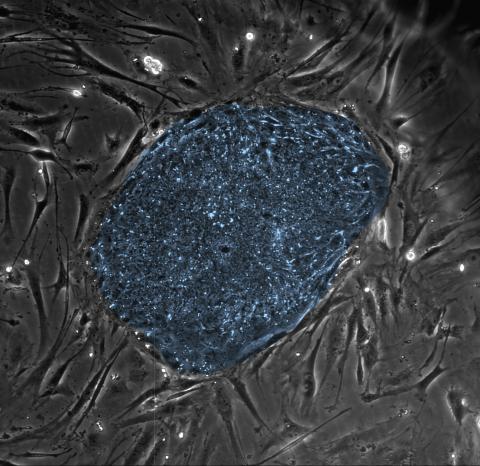
2608: Human embryonic stem cells
2608: Human embryonic stem cells
The center cluster of cells, colored blue, shows a colony of human embryonic stem cells. These cells, which arise at the earliest stages of development, are capable of differentiating into any of the 220 types of cells in the human body and can provide access to cells for basic research and potential therapies. This image is from the lab of the University of Wisconsin-Madison's James Thomson.
James Thomson, University of Wisconsin-Madison
View Media
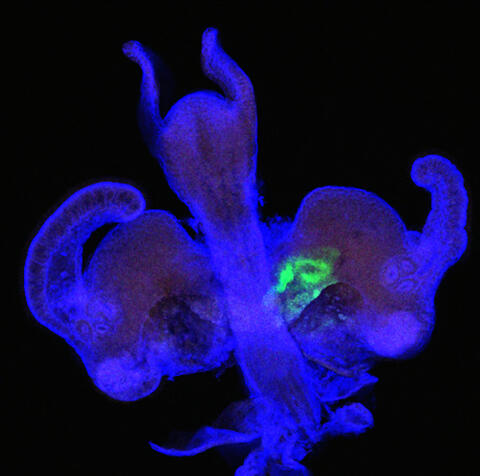
7020: Bacterial symbionts colonizing the crypts of a juvenile Hawaiian bobtail squid light organ
7020: Bacterial symbionts colonizing the crypts of a juvenile Hawaiian bobtail squid light organ
A light organ (~0.5 mm across) of a Hawaiian bobtail squid, Euprymna scolopes, stained blue. At the time of this image, the crypts within the tissues of only one side of the organ had been colonized by green-fluorescent protein-labeled Vibrio fischeri cells, which can be seen here in green. This image was taken using confocal fluorescence microscopy.
Related to images 7016, 7017, 7018, and 7019.
Related to images 7016, 7017, 7018, and 7019.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
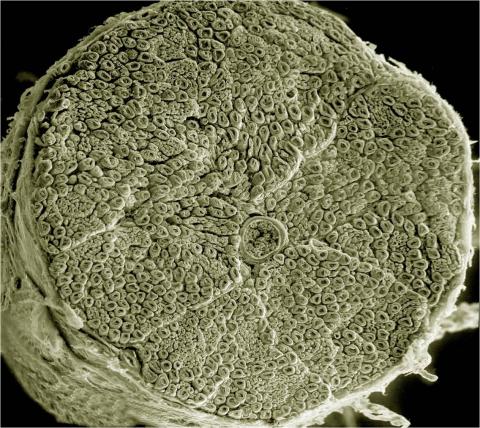
3387: NCMIR human spinal nerve
3387: NCMIR human spinal nerve
Spinal nerves are part of the peripheral nervous system. They run within the spinal column to carry nerve signals to and from all parts of the body. The spinal nerves enable all the movements we do, from turning our heads to wiggling our toes, control the movements of our internal organs, such as the colon and the bladder, as well as allow us to feel touch and the location of our limbs.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media
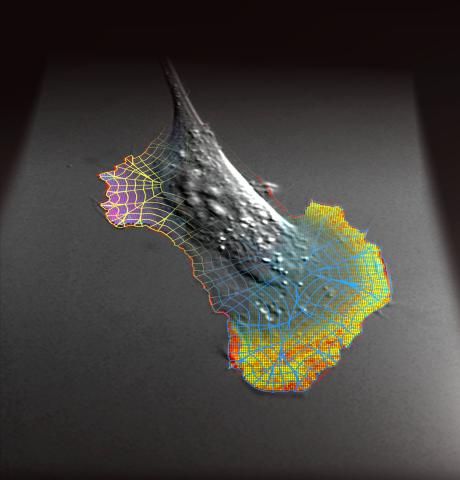
2802: Biosensors illustration
2802: Biosensors illustration
A rendering of an activity biosensor image overlaid with a cell-centered frame of reference used for image analysis of signal transduction. This is an example of NIH-supported research on single-cell analysis. Related to 2798 , 2799, 2800, 2801 and 2803.
Gaudenz Danuser, Harvard Medical School
View Media
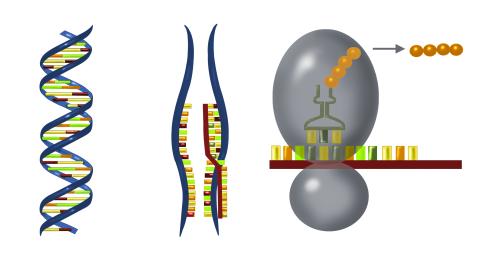
2547: Central dogma, illustrated
2547: Central dogma, illustrated
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a labeled version of this illustration and 2549 for a labeled and numbered version. Featured in The New Genetics.
Crabtree + Company
View Media
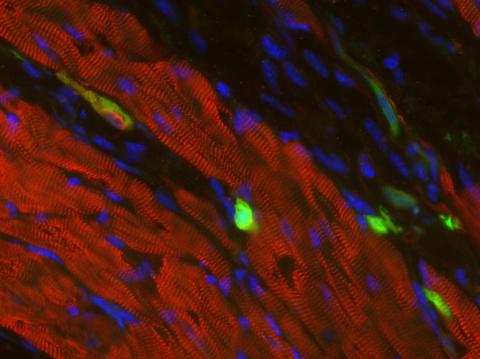
3273: Heart muscle with reprogrammed skin cells
3273: Heart muscle with reprogrammed skin cells
Skins cells were reprogrammed into heart muscle cells. The cells highlighted in green are remaining skin cells. Red indicates a protein that is unique to heart muscle. The technique used to reprogram the skin cells into heart cells could one day be used to mend heart muscle damaged by disease or heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Deepak Srivastava, Gladstone Institute of Cardiovascular Disease, via CIRM
View Media
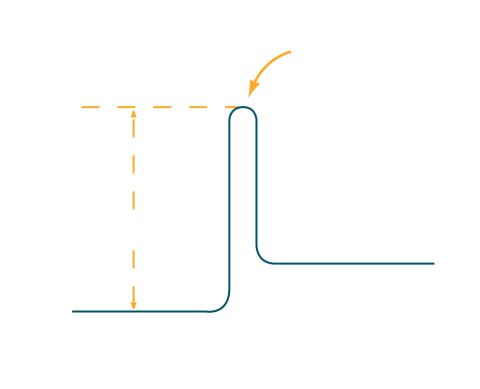
2525: Activation energy
2525: Activation energy
To become products, reactants must overcome an energy hill. See image 2526 for a labeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
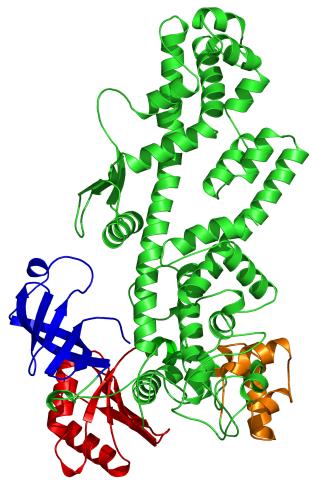
2338: Tex protein
2338: Tex protein
Model of a member from the Tex protein family, which is implicated in transcriptional regulation and highly conserved in eukaryotes and prokaryotes. The structure shows significant homology to a human transcription elongation factor that may regulate multiple steps in mRNA synthesis.
New York Structural GenomiX Research Consortium, PSI
View Media
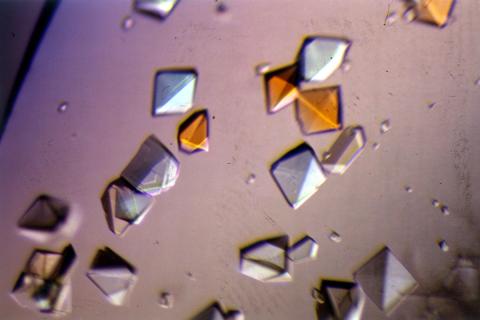
2412: Pig alpha amylase
2412: Pig alpha amylase
Crystals of porcine alpha amylase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
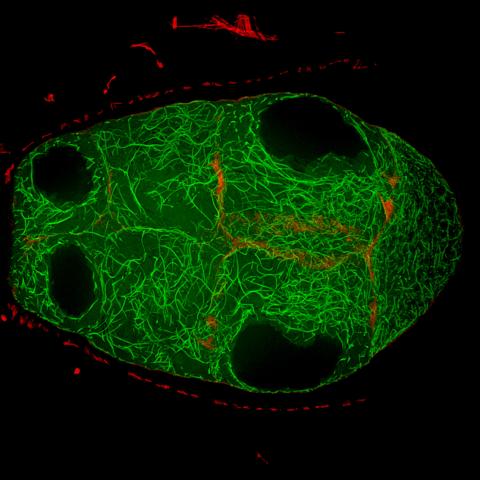
6811: Fruit fly egg chamber
6811: Fruit fly egg chamber
A fruit fly (Drosophila melanogaster) egg chamber with microtubules shown in green and actin filaments shown in red. Egg chambers are multicellular structures in fruit flies ovaries that each give rise to a single egg. Microtubules and actin filaments give the chambers structure and shape. This image was captured using a confocal microscope.
More information on the research that produced this image can be found in the Current Biology paper "Gatekeeper function for Short stop at the ring canals of the Drosophila ovary" by Lu et al.
More information on the research that produced this image can be found in the Current Biology paper "Gatekeeper function for Short stop at the ring canals of the Drosophila ovary" by Lu et al.
Vladimir I. Gelfand, Feinberg School of Medicine, Northwestern University.
View Media
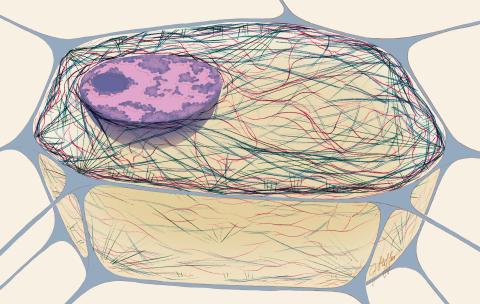
1272: Cytoskeleton
1272: Cytoskeleton
The three fibers of the cytoskeleton--microtubules in blue, intermediate filaments in red, and actin in green--play countless roles in the cell.
Judith Stoffer
View Media
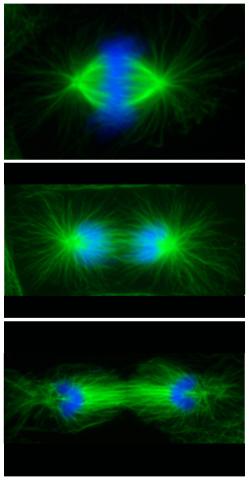
3442: Cell division phases in Xenopus frog cells
3442: Cell division phases in Xenopus frog cells
These images show three stages of cell division in Xenopus XL177 cells, which are derived from tadpole epithelial cells. They are (from top): metaphase, anaphase and telophase. The microtubules are green and the chromosomes are blue. Related to 3443.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison
View Media
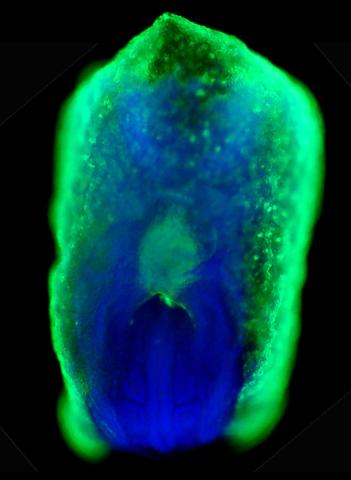
2607: Mouse embryo showing Smad4 protein
2607: Mouse embryo showing Smad4 protein
This eerily glowing blob isn't an alien or a creature from the deep sea--it's a mouse embryo just eight and a half days old. The green shell and core show a protein called Smad4. In the center, Smad4 is telling certain cells to begin forming the mouse's liver and pancreas. Researchers identified a trio of signaling pathways that help switch on Smad4-making genes, starting immature cells on the path to becoming organs. The research could help biologists learn how to grow human liver and pancreas tissue for research, drug testing and regenerative medicine. In addition to NIGMS, NIH's National Institute of Diabetes and Digestive and Kidney Diseases also supported this work.
Kenneth Zaret, Fox Chase Cancer Center
View Media
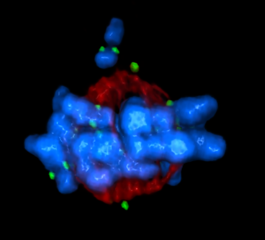
5765: Mitotic cell awaits chromosome alignment
5765: Mitotic cell awaits chromosome alignment
During mitosis, spindle microtubules (red) attach to chromosome pairs (blue), directing them to the spindle equator. This midline alignment is critical for equal distribution of chromosomes in the dividing cell. Scientists are interested in how the protein kinase Plk1 (green) regulates this activity in human cells. Image is a volume projection of multiple deconvolved z-planes acquired with a Nikon widefield fluorescence microscope. This image was chosen as a winner of the 2016 NIH-funded research image call. Related to image 5766.
The research that led to this image was funded by NIGMS.
View Media
The research that led to this image was funded by NIGMS.
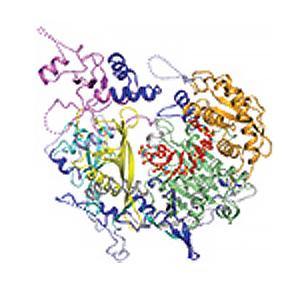
3408: Kluyveromyces polysporus Argonaute bound to guide RNA
3408: Kluyveromyces polysporus Argonaute bound to guide RNA
A segment of siRNA, shown in red, guides a "slicer" protein called Argonaute (multi-colored twists and corkscrews) to the target RNA molecules.
Kotaro Nakanishi and David Weinberg, Massachusetts Institute of Technology
View Media
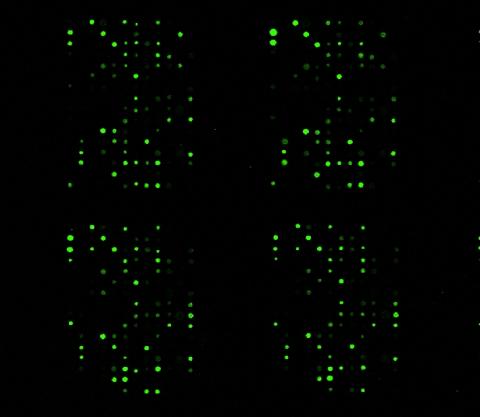
1265: Glycan arrays
1265: Glycan arrays
The signal is obtained by allowing proteins in human serum to interact with glycan (polysaccharide) arrays. The arrays are shown in replicate so the pattern is clear. Each spot contains a specific type of glycan. Proteins have bound to the spots highlighted in green.
Ola Blixt, Scripps Research Institute
View Media

1271: Cone cell
1271: Cone cell
The cone cell of the eye allows you to see in color. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media
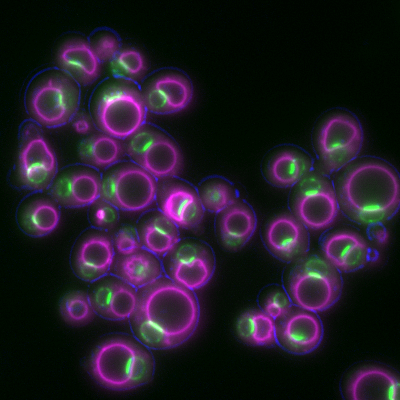
6772: Yeast cells responding to a glucose shortage
6772: Yeast cells responding to a glucose shortage
These yeast cells were exposed to a glucose (sugar) shortage. This caused the cells to compartmentalize HMGCR (green)—an enzyme involved in making cholesterol—to a patch on the nuclear envelope next to the vacuole/lysosome (purple). This process enhanced HMGCR activity and helped the yeast adapt to the glucose shortage. Researchers hope that understanding how yeast regulate cholesterol could ultimately lead to new ways to treat high cholesterol in people. This image was captured using a fluorescence microscope.
Mike Henne, University of Texas Southwestern Medical Center.
View Media
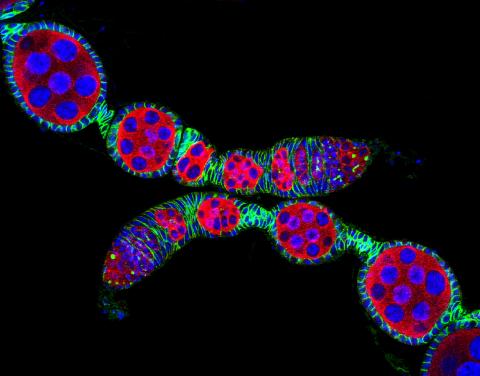
5772: Confocal microscopy image of two Drosophila ovarioles
5772: Confocal microscopy image of two Drosophila ovarioles
Ovarioles in female insects are tubes in which egg cells (called oocytes) form at one end and complete their development as they reach the other end of the tube. This image, taken with a confocal microscope, shows ovarioles in a very popular lab animal, the fruit fly Drosophila. The basic structure of ovarioles supports very rapid egg production, with some insects (like termites) producing several thousand eggs per day. Each insect ovary typically contains four to eight ovarioles, but this number varies widely depending on the insect species.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
Scientists use insect ovarioles, for example, to study the basic processes that help various insects, including those that cause disease (like some mosquitos and biting flies), reproduce very quickly.
2004 Olympus BioScapes Competition
View Media
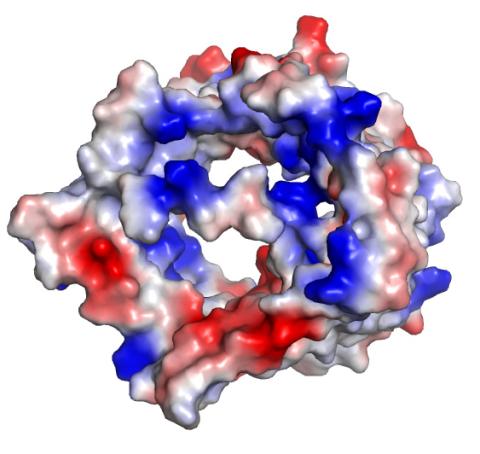
2491: VDAC-1 (2)
2491: VDAC-1 (2)
The structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to images 2494, 2495, and 2488.
Related to images 2494, 2495, and 2488.
Gerhard Wagner, Harvard Medical School
View Media
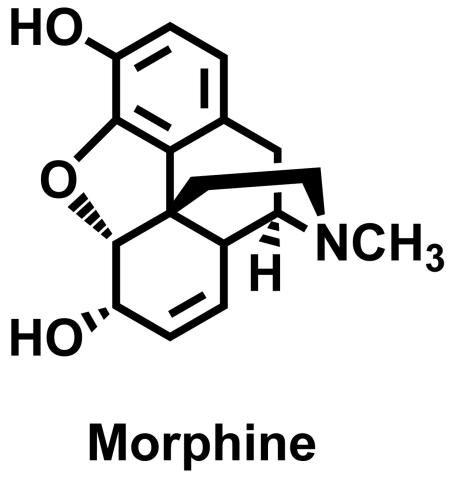
3438: Morphine Structure
3438: Morphine Structure
The chemical structure of the morphine molecule
Judy Coyle, Donald Danforth Plant Science Center
View Media
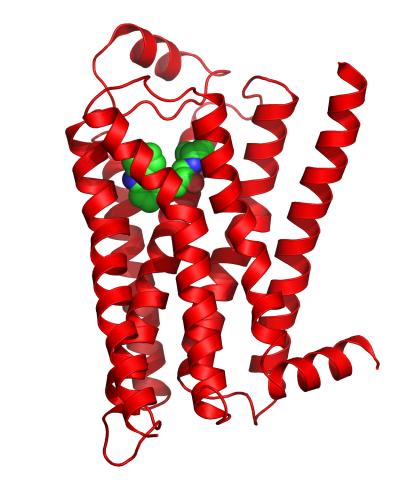
3358: Beta 2-adrenergic receptor
3358: Beta 2-adrenergic receptor
The receptor is shown bound to a partial inverse agonist, carazolol.
Raymond Stevens, The Scripps Research Institute
View Media

2407: Jack bean concanavalin A
2407: Jack bean concanavalin A
Crystals of jack bean concanavalin A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

2443: Mapping human genetic variation
2443: Mapping human genetic variation
This map paints a colorful portrait of human genetic variation around the world. Researchers analyzed the DNA of 485 people and tinted the genetic types in different colors to produce one of the most detailed maps of its kind ever made. The map shows that genetic variation decreases with increasing distance from Africa, which supports the idea that humans originated in Africa, spread to the Middle East, then to Asia and Europe, and finally to the Americas. The data also offers a rich resource that scientists could use to pinpoint the genetic basis of diseases prevalent in diverse populations. Featured in the March 19, 2008, issue of Biomedical Beat.
Noah Rosenberg and Martin Soave, University of Michigan
View Media

3424: White Poppy
3424: White Poppy
A white poppy. View cropped image of a poppy here 3423.
Judy Coyle, Donald Danforth Plant Science Center
View Media
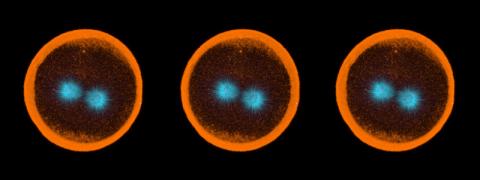
1050: Sea urchin embryo 04
1050: Sea urchin embryo 04
Stereo triplet of a sea urchin embryo stained to reveal actin filaments (orange) and microtubules (blue). This image is part of a series of images: image 1047, image 1048, image 1049, image 1051 and image 1052.
George von Dassow, University of Washington
View Media
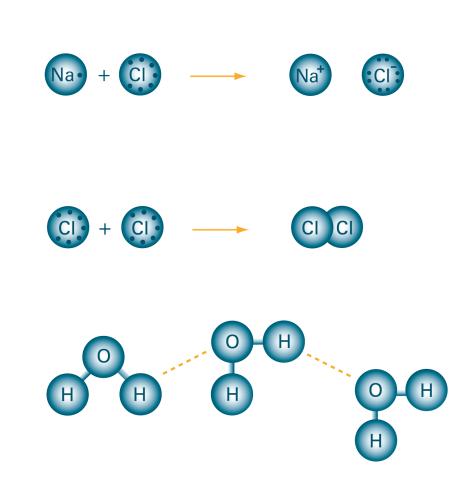
2519: Bond types
2519: Bond types
Ionic and covalent bonds hold molecules, like sodium chloride and chlorine gas, together. Hydrogen bonds among molecules, notably involving water, also play an important role in biology. See image 2520 for a labeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media
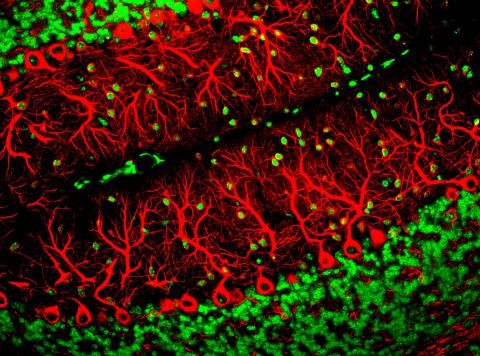
3637: Purkinje cells are one of the main cell types in the brain
3637: Purkinje cells are one of the main cell types in the brain
This image captures Purkinje cells (red), one of the main types of nerve cell found in the brain. These cells have elaborate branching structures called dendrites that receive signals from other nerve cells.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Yinghua Ma and Timothy Vartanian, Cornell University, Ithaca, N.Y.
View Media
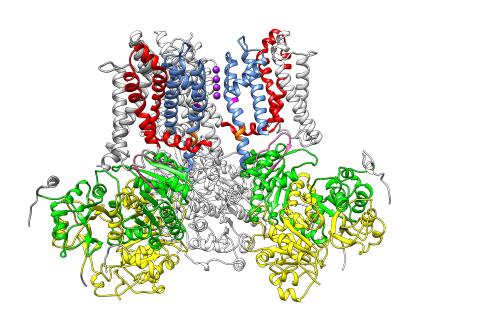
3487: Ion channel
3487: Ion channel
A special "messy" region of a potassium ion channel is important in its function.
Yu Zhoi, Christopher Lingle Laboratory, Washington University School of Medicine in St. Louis
View Media
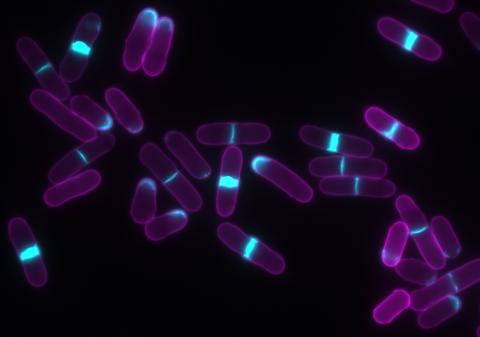
6797: Yeast cells with accumulated cell wall material
6797: Yeast cells with accumulated cell wall material
Yeast cells that abnormally accumulate cell wall material (blue) at their ends and, when preparing to divide, in their middles. This image was captured using wide-field microscopy with deconvolution.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Related to images 6791, 6792, 6793, 6794, 6798, and videos 6795 and 6796.
Alaina Willet, Kathy Gould’s lab, Vanderbilt University.
View Media
3597: DNA replication origin recognition complex (ORC)
3597: DNA replication origin recognition complex (ORC)
A study published in March 2012 used cryo-electron microscopy to determine the structure of the DNA replication origin recognition complex (ORC), a semi-circular, protein complex (yellow) that recognizes and binds DNA to start the replication process. The ORC appears to wrap around and bend approximately 70 base pairs of double stranded DNA (red and blue). Also shown is the protein Cdc6 (green), which is also involved in the initiation of DNA replication. Related to video 3307 that shows the structure from different angles. From a Brookhaven National Laboratory news release, "Study Reveals How Protein Machinery Binds and Wraps DNA to Start Replication."
Huilin Li, Brookhaven National Laboratory
View Media

5855: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 1
5855: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 1
Superresolution microscopy work on endoplasmic reticulum (ER) in the peripheral areas of the cell showing details of the structure and arrangement in a complex web of tubes. The ER is a continuous membrane that extends like a net from the envelope of the nucleus outward to the cell membrane. The ER plays several roles within the cell, such as in protein and lipid synthesis and transport of materials between organelles. The ER has a flexible structure to allow it to accomplish these tasks by changing shape as conditions in the cell change. Shown here an image created by super-resolution microscopy of the ER in the peripheral areas of the cell showing details of the structure and the arrangements in a complex web of tubes. Related to images 5856 and 5857.
Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia
View Media
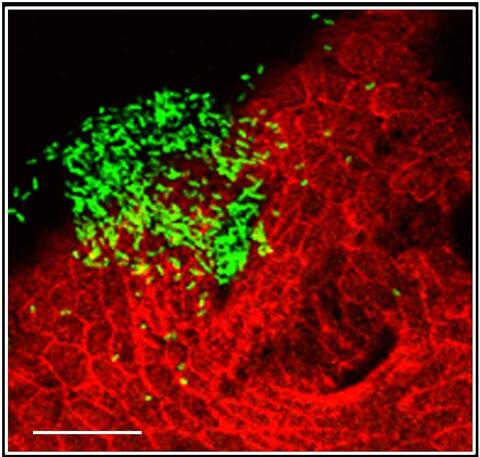
7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
7019: Bacterial cells aggregated above a light-organ pore of the Hawaiian bobtail squid
The beating of cilia on the outside of the Hawaiian bobtail squid’s light organ concentrates Vibrio fischeri cells (green) present in the seawater into aggregates near the pore-containing tissue (red). From there, the bacterial cells (~2 mm) swim to the pores and migrate through a bottleneck into the interior crypts where a population of symbionts grow and remain for the life of the host. This image was taken using confocal fluorescence microscopy.
Related to images 7016, 7017, 7018, and 7020.
Related to images 7016, 7017, 7018, and 7020.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media
