Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
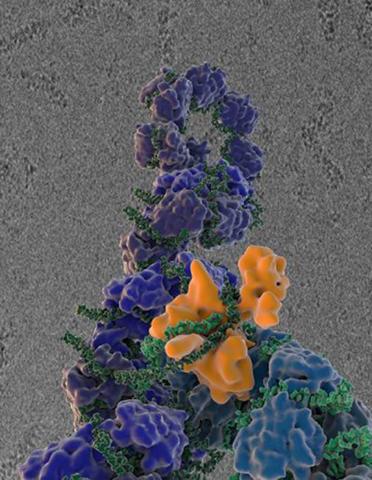
3434: Flu virus proteins during self-replication
3434: Flu virus proteins during self-replication
Influenza (flu) virus proteins in the act of self-replication. Viral nucleoprotein (blue) encapsidates [encapsulates] the RNA genome (green). The influenza virus polymerase (orange) reads and copies the RNA genome. In the background is an image of influenza virus ribonucleoprotein complexes observed using cryo-electron microscopy. This image is from a November 2012 News Release.
Scripps Research Institute in La Jolla, CA
View Media
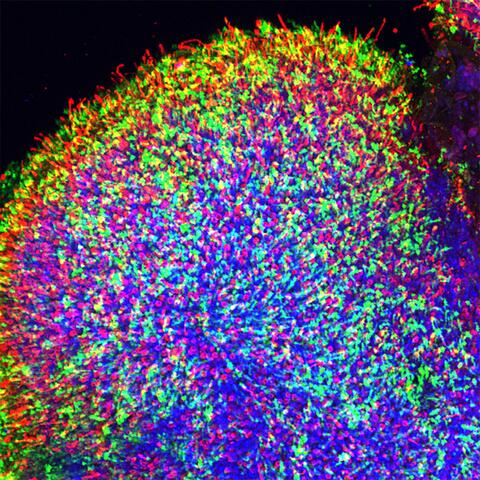
6748: Human retinal organoid
6748: Human retinal organoid
A replica of a human retina grown from stem cells. It shows rod photoreceptors (nerve cells responsible for dark vision) in green and red/green cones (nerve cells responsible for red and green color vision) in red. The cell nuclei are stained blue. This image was captured using a confocal microscope.
Kevin Eliceiri, University of Wisconsin-Madison.
View Media
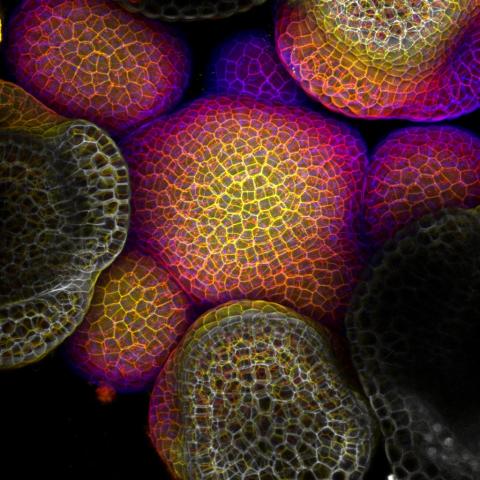
3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
In plants, as in animals, stem cells can transform into a variety of different cell types. The stem cells at the growing tip of this Arabidopsis plant will soon become flowers. Arabidopsis is frequently studied by cellular and molecular biologists because it grows rapidly (its entire life cycle is only 6 weeks), produces lots of seeds, and has a genome that is easy to manipulate.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Arun Sampathkumar and Elliot Meyerowitz, California Institute of Technology
View Media
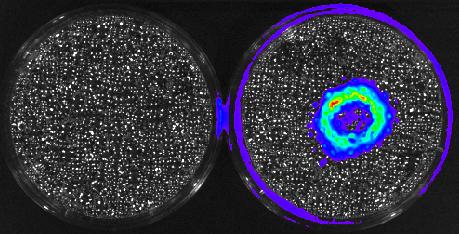
3480: Cancer Cells Glowing from Luciferin
3480: Cancer Cells Glowing from Luciferin
The activator cancer cell culture, right, contains a chemical that causes the cells to emit light when in the presence of immune cells.
Mark Sellmyer, Stanford University School of Medicine
View Media
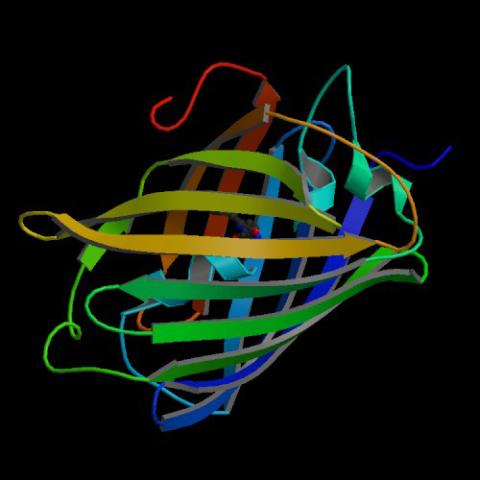
2317: Fruitful dyes
2317: Fruitful dyes
These colorful, computer-generated ribbons show the backbone of a molecule that glows a fluorescent red. The molecule, called mStrawberry, was created by chemists based on a protein found in the ruddy lips of a coral. Scientists use the synthetic molecule and other "fruity" ones like it as a dye to mark and study cell structures.
Roger Y. Tsien, University of California, San Diego
View Media
1337: Bicycling cell
1337: Bicycling cell
A humorous treatment of the concept of a cycling cell.
Judith Stoffer
View Media
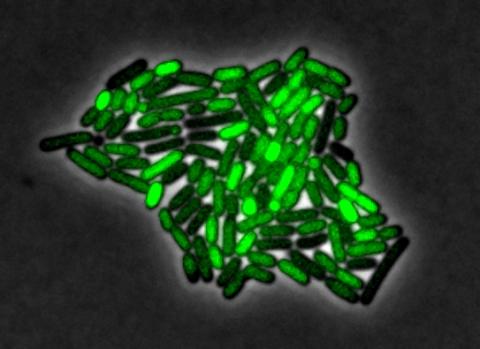
3253: Pulsating response to stress in bacteria
3253: Pulsating response to stress in bacteria
By attaching fluorescent proteins to the genetic circuit responsible for B. subtilis's stress response, researchers can observe the cells' pulses as green flashes. In response to a stressful environment like one lacking food, B. subtilis activates a large set of genes that help it respond to the hardship. Instead of leaving those genes on as previously thought, researchers discovered that the bacteria flip the genes on and off, increasing the frequency of these pulses with increasing stress. See entry 3254 for the related video.
Michael Elowitz, Caltech University
View Media
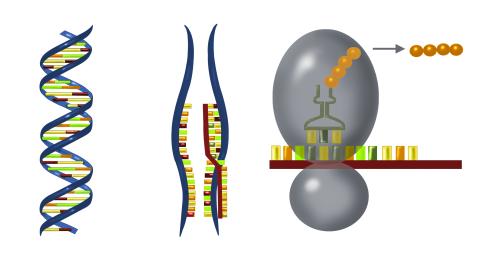
2547: Central dogma, illustrated
2547: Central dogma, illustrated
DNA encodes RNA, which encodes protein. DNA is transcribed to make messenger RNA (mRNA). The mRNA sequence (dark red strand) is complementary to the DNA sequence (blue strand). On ribosomes, transfer RNA (tRNA) reads three nucleotides at a time in mRNA to bring together the amino acids that link up to make a protein. See image 2548 for a labeled version of this illustration and 2549 for a labeled and numbered version. Featured in The New Genetics.
Crabtree + Company
View Media

2443: Mapping human genetic variation
2443: Mapping human genetic variation
This map paints a colorful portrait of human genetic variation around the world. Researchers analyzed the DNA of 485 people and tinted the genetic types in different colors to produce one of the most detailed maps of its kind ever made. The map shows that genetic variation decreases with increasing distance from Africa, which supports the idea that humans originated in Africa, spread to the Middle East, then to Asia and Europe, and finally to the Americas. The data also offers a rich resource that scientists could use to pinpoint the genetic basis of diseases prevalent in diverse populations. Featured in the March 19, 2008, issue of Biomedical Beat.
Noah Rosenberg and Martin Soave, University of Michigan
View Media
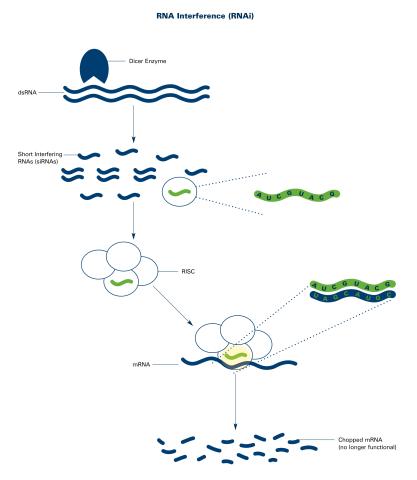
2559: RNA interference (with labels)
2559: RNA interference (with labels)
RNA interference or RNAi is a gene-silencing process in which double-stranded RNAs trigger the destruction of specific RNAs. See 2558 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
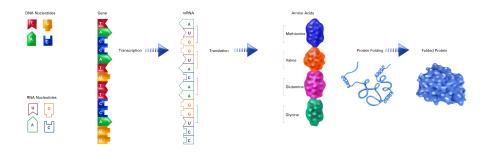
2510: From DNA to Protein (labeled)
2510: From DNA to Protein (labeled)
The genetic code in DNA is transcribed into RNA, which is translated into proteins with specific sequences. During transcription, nucleotides in DNA are copied into RNA, where they are read three at a time to encode the amino acids in a protein. Many parts of a protein fold as the amino acids are strung together.
See image 2509 for an unlabeled version of this illustration.
Featured in The Structures of Life.
See image 2509 for an unlabeled version of this illustration.
Featured in The Structures of Life.
Crabtree + Company
View Media

3272: Ear hair cells derived from embryonic stem cells
3272: Ear hair cells derived from embryonic stem cells
Mouse embryonic stem cells matured into this bundle of hair cells similar to the ones that transmit sound in the ear. These cells could one day be transplanted as a therapy for some forms of deafness, or they could be used to screen drugs to treat deafness. The hairs are shown at 23,000 times magnification via scanning electron microscopy. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Stefen Heller, Stanford University, via CIRM
View Media
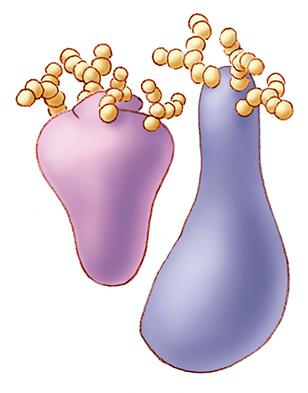
1270: Glycoproteins
1270: Glycoproteins
About half of all human proteins include chains of sugar molecules that are critical for the proteins to function properly. Appears in the NIGMS booklet Inside the Cell.
Judith Stoffer
View Media
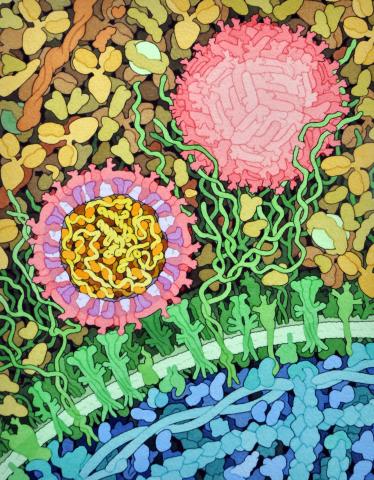
6998: Zika virus
6998: Zika virus
Zika virus is shown in cross section at center left. On the outside, it includes envelope protein (red) and membrane protein (magenta) embedded in a lipid membrane (light purple). Inside, the RNA genome (yellow) is associated with capsid proteins (orange). The viruses are shown interacting with receptors on the cell surface (green) and are surrounded by blood plasma molecules at the top.
Amy Wu and Christine Zardecki, RCSB Protein Data Bank.
View Media
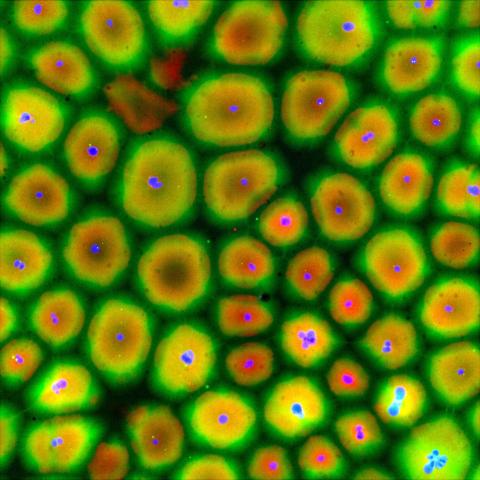
6584: Cell-like compartments from frog eggs
6584: Cell-like compartments from frog eggs
Cell-like compartments that spontaneously emerged from scrambled frog eggs, with nuclei (blue) from frog sperm. Endoplasmic reticulum (red) and microtubules (green) are also visible. Image created using epifluorescence microscopy.
For more photos of cell-like compartments from frog eggs view: 6585, 6586, 6591, 6592, and 6593.
For videos of cell-like compartments from frog eggs view: 6587, 6588, 6589, and 6590.
Xianrui Cheng, Stanford University School of Medicine.
View Media
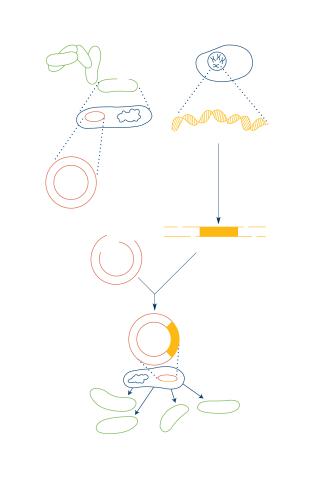
2564: Recombinant DNA
2564: Recombinant DNA
To splice a human gene into a plasmid, scientists take the plasmid out of an E. coli bacterium, cut the plasmid with a restriction enzyme, and splice in human DNA. The resulting hybrid plasmid can be inserted into another E. coli bacterium, where it multiplies along with the bacterium. There, it can produce large quantities of human protein. See image 2565 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
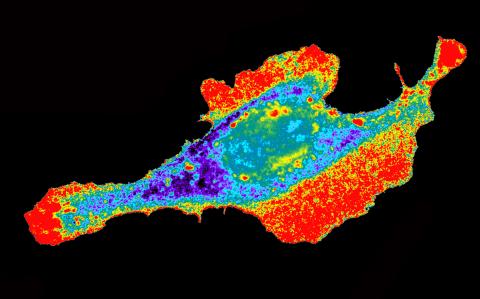
2453: Seeing signaling protein activation in cells 03
2453: Seeing signaling protein activation in cells 03
Cdc42, a member of the Rho family of small guanosine triphosphatase (GTPase) proteins, regulates multiple cell functions, including motility, proliferation, apoptosis, and cell morphology. In order to fulfill these diverse roles, the timing and location of Cdc42 activation must be tightly controlled. Klaus Hahn and his research group use special dyes designed to report protein conformational changes and interactions, here in living neutrophil cells. Warmer colors in this image indicate higher levels of activation. Cdc42 looks to be activated at cell protrusions.
Related to images 2451, 2452, and 2454.
Related to images 2451, 2452, and 2454.
Klaus Hahn, University of North Carolina, Chapel Hill Medical School
View Media

7014: Flagellated bacterial cells
7014: Flagellated bacterial cells
Vibrio fischeri (2 mm in length) is the exclusive symbiotic partner of the Hawaiian bobtail squid, Euprymna scolopes. After this bacterium uses its flagella to swim from the seawater into the light organ of a newly hatched juvenile, it colonizes the host and loses the appendages. This image was taken using a scanning electron microscope.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media

1311: Housekeeping cell illustration
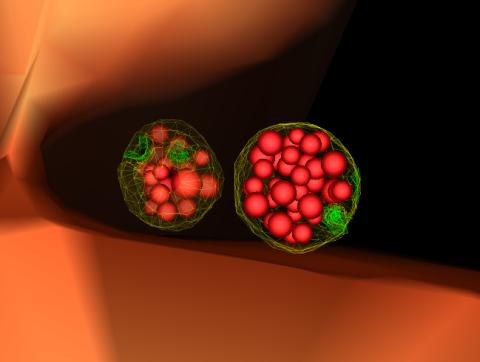
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
5767: Multivesicular bodies containing intralumenal vesicles assemble at the vacuole 3
Collecting and transporting cellular waste and sorting it into recylable and nonrecylable pieces is a complex business in the cell. One key player in that process is the endosome, which helps collect, sort and transport worn-out or leftover proteins with the help of a protein assembly called the endosomal sorting complexes for transport (or ESCRT for short). These complexes help package proteins marked for breakdown into intralumenal vesicles, which, in turn, are enclosed in multivesicular bodies for transport to the places where the proteins are recycled or dumped. In this image, two multivesicular bodies (with yellow membranes) contain tiny intralumenal vesicles (with a diameter of only 25 nanometers; shown in red) adjacent to the cell's vacuole (in orange).
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Scientists working with baker's yeast (Saccharomyces cerevisiae) study the budding inward of the limiting membrane (green lines on top of the yellow lines) into the intralumenal vesicles. This tomogram was shot with a Tecnai F-20 high-energy electron microscope, at 29,000x magnification, with a 0.7-nm pixel, ~4-nm resolution.
To learn more about endosomes, see the Biomedical Beat blog post The Cell’s Mailroom. Related to a microscopy photograph 5768 that was used to generate this illustration and a zoomed-out version 5769 of this illustration.
Matthew West and Greg Odorizzi, University of Colorado
View Media
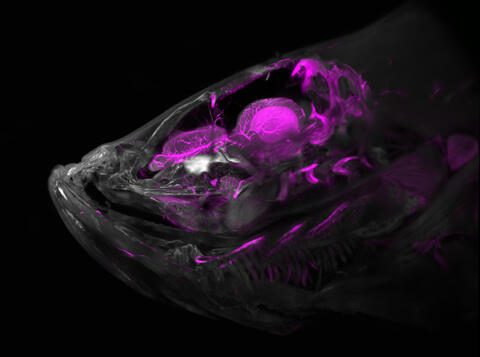
6934: Zebrafish head vasculature
6934: Zebrafish head vasculature
A zebrafish head with blood vessels shown in purple. Researchers often study zebrafish because they share many genes with humans, grow and reproduce quickly, and have see-through eggs and embryos, which make it easy to study early stages of development.
This image was captured using a light sheet microscope.
Related to video 6933.
This image was captured using a light sheet microscope.
Related to video 6933.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media
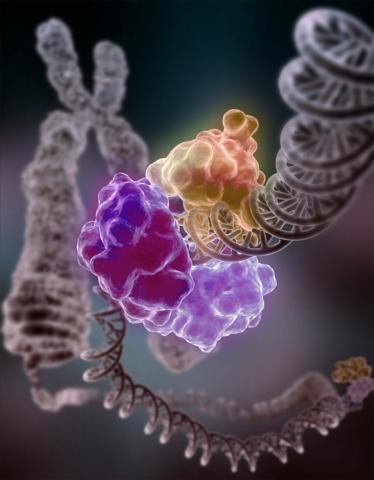
3493: Repairing DNA
3493: Repairing DNA
Like a watch wrapped around a wrist, a special enzyme encircles the double helix to repair a broken strand of DNA. Without molecules that can mend such breaks, cells can malfunction, die, or become cancerous. Related to image 2330.
Tom Ellenberger, Washington University School of Medicine
View Media
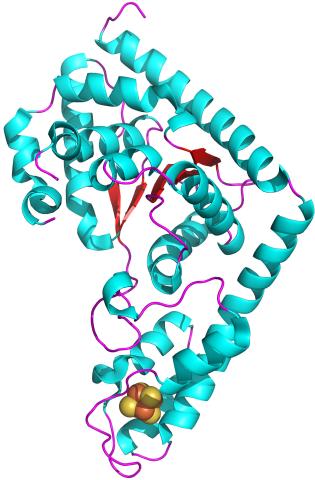
2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
2483: Trp_RS - tryptophanyl tRNA-synthetase family of enzymes
This image represents the structure of TrpRS, a novel member of the tryptophanyl tRNA-synthetase family of enzymes. By helping to link the amino acid tryptophan to a tRNA molecule, TrpRS primes the amino acid for use in protein synthesis. A cluster of iron and sulfur atoms (orange and red spheres) was unexpectedly found in the anti-codon domain, a key part of the molecule, and appears to be critical for the function of the enzyme. TrpRS was discovered in Thermotoga maritima, a rod-shaped bacterium that flourishes in high temperatures.
View Media
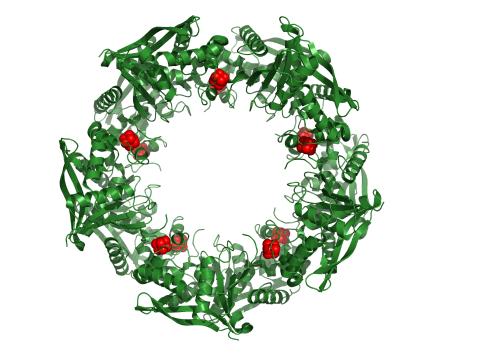
3720: Cas4 nuclease protein structure
3720: Cas4 nuclease protein structure
This wreath represents the molecular structure of a protein, Cas4, which is part of a system, known as CRISPR, that bacteria use to protect themselves against viral invaders. The green ribbons show the protein's structure, and the red balls show the location of iron and sulfur molecules important for the protein's function. Scientists harnessed Cas9, a different protein in the bacterial CRISPR system, to create a gene-editing tool known as CRISPR-Cas9. Using this tool, researchers are able to study a range of cellular processes and human diseases more easily, cheaply and precisely. In December, 2015, Science magazine recognized the CRISPR-Cas9 gene-editing tool as the "breakthrough of the year." Read more about Cas4 in the December 2015 Biomedical Beat post A Holiday-Themed Image Collection.
Fred Dyda, NIDDK
View Media
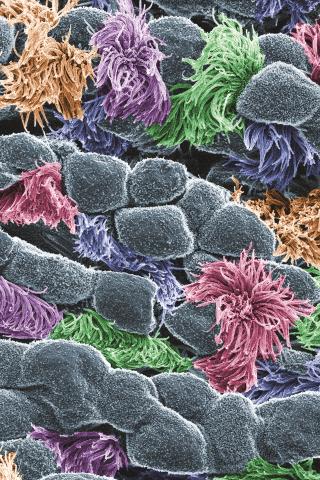
3646: Cells lining the trachea
3646: Cells lining the trachea
In this image, viewed with a ZEISS ORION NanoFab microscope, the community of cells lining a mouse airway is magnified more than 10,000 times. This collection of cells, known as the mucociliary escalator, is also found in humans. It is our first line of defense against inhaled bacteria, allergens, pollutants, and debris. Malfunctions in the system can cause or aggravate lung infections and conditions such as asthma and chronic obstructive pulmonary disease. The cells shown in gray secrete mucus, which traps inhaled particles. The colored cells sweep the mucus layer out of the lungs.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Eva Mutunga and Kate Klein, University of the District of Columbia and National Institute of Standards and Technology
View Media

1313: Cell eyes clock
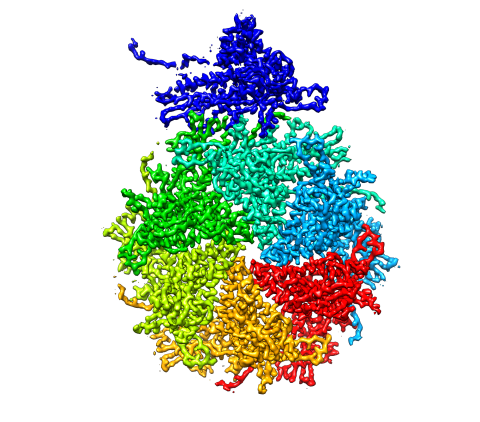
5875: Bacteriophage P22 capsid, detail
5875: Bacteriophage P22 capsid, detail
Detail of a subunit of the capsid, or outer cover, of bacteriophage P22, a virus that infects the Salmonella bacteria. Cryo-electron microscopy (cryo-EM) was used to capture details of the capsid proteins, each shown here in a separate color. Thousands of cryo-EM scans capture the structure and shape of all the individual proteins in the capsid and their position relative to other proteins. A computer model combines these scans into the image shown here. Related to image 5874.
Dr. Wah Chiu, Baylor College of Medicine
View Media
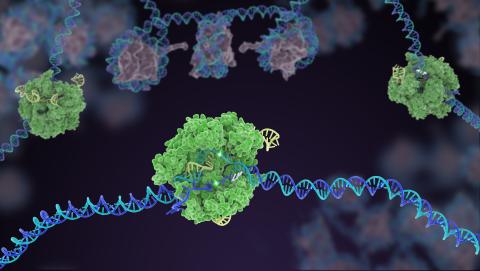
5816: Cas9 protein involved in the CRISPR gene-editing technology
5816: Cas9 protein involved in the CRISPR gene-editing technology
In the gene-editing tool CRISPR, a small strand of RNA identifies a specific chunk of DNA. Then the enzyme Cas9 (green) swoops in and cuts the double-stranded DNA (blue/purple) in two places, removing the specific chunk.
Janet Iwasa
View Media

1089: Natcher Building 09
1089: Natcher Building 09
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media
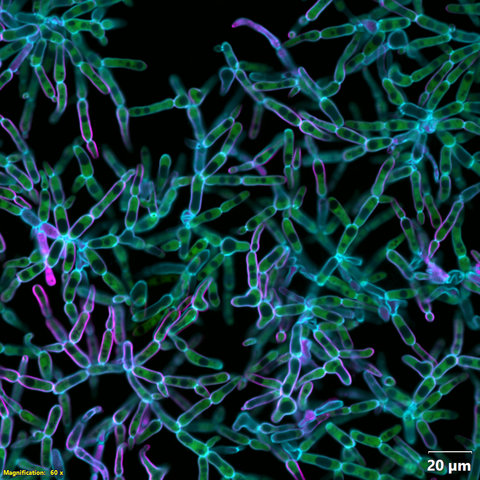
6970: Snowflake yeast 2
6970: Snowflake yeast 2
Multicellular yeast called snowflake yeast that researchers created through many generations of directed evolution from unicellular yeast. Cells are connected to one another by their cell walls, shown in blue. Stained cytoplasm (green) and membranes (magenta) show that the individual cells remain separate. This image was captured using spinning disk confocal microscopy.
Related to images 6969 and 6971.
Related to images 6969 and 6971.
William Ratcliff, Georgia Institute of Technology.
View Media

2395: Fungal lipase (1)
2395: Fungal lipase (1)
Crystals of fungal lipase protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
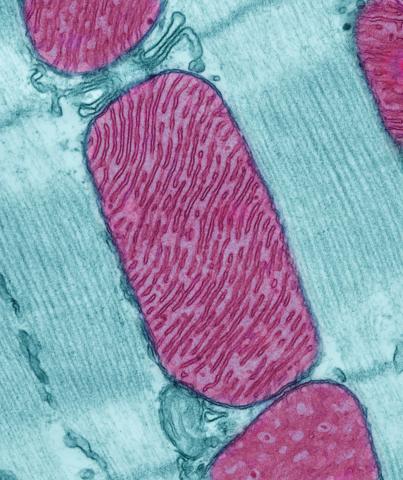
3661: Mitochondria from rat heart muscle cell
3661: Mitochondria from rat heart muscle cell
These mitochondria (red) are from the heart muscle cell of a rat. Mitochondria have an inner membrane that folds in many places (and that appears here as striations). This folding vastly increases the surface area for energy production. Nearly all our cells have mitochondria. Related to image 3664.
National Center for Microscopy and Imaging Research
View Media
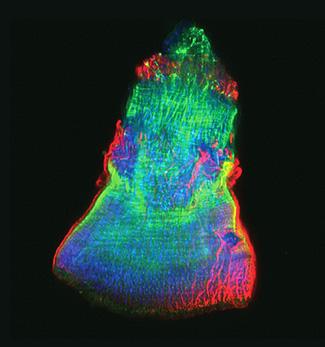
3593: Isolated Planarian Pharynx
3593: Isolated Planarian Pharynx
The feeding tube, or pharynx, of a planarian worm with cilia shown in red and muscle fibers shown in green
View Media
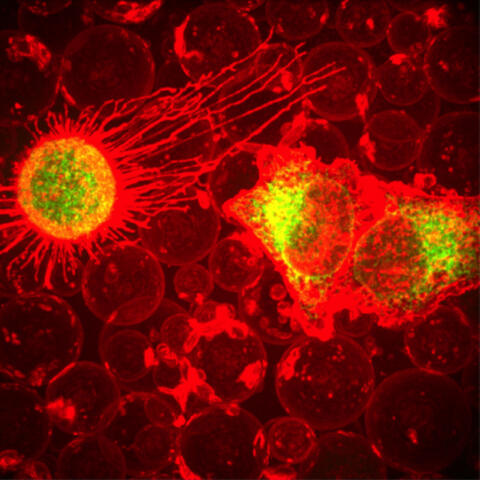
5887: Plasma-Derived Membrane Vesicles
5887: Plasma-Derived Membrane Vesicles
This fiery image doesn’t come from inside a bubbling volcano. Instead, it shows animal cells caught in the act of making bubbles, or blebbing. Some cells regularly pinch off parts of their membranes to produce bubbles filled with a mix of proteins and fats. The bubbles (red) are called plasma-derived membrane vesicles, or PMVs, and can travel to other parts of the body where they may aid in cell-cell communication. The University of Texas, Austin, researchers responsible for this photo are exploring ways to use PMVs to deliver medicines to precise locations in the body.
This image, entered in the Biophysical Society’s 2017 Art of Science Image contest, used two-channel spinning disk confocal fluorescence microscopy. It was also featured in the NIH Director’s Blog in May 2017.
This image, entered in the Biophysical Society’s 2017 Art of Science Image contest, used two-channel spinning disk confocal fluorescence microscopy. It was also featured in the NIH Director’s Blog in May 2017.
Jeanne Stachowiak, University of Texas at Austin
View Media
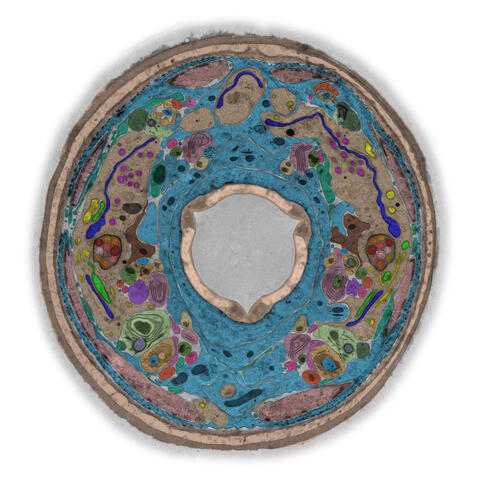
5759: TEM cross-section of C. elegans (roundworm)
5759: TEM cross-section of C. elegans (roundworm)
The worm Caenorhabditis elegans is a popular laboratory animal because its small size and fairly simple body make it easy to study. Scientists use this small worm to answer many research questions in developmental biology, neurobiology, and genetics. This image, which was taken with transmission electron microscopy (TEM), shows a cross-section through C. elegans, revealing various internal structures.
The image is from a figure in an article published in the journal eLife. There is an annotated version of this graphic at 5760.
The image is from a figure in an article published in the journal eLife. There is an annotated version of this graphic at 5760.
Piali Sengupta, Brandeis University
View Media
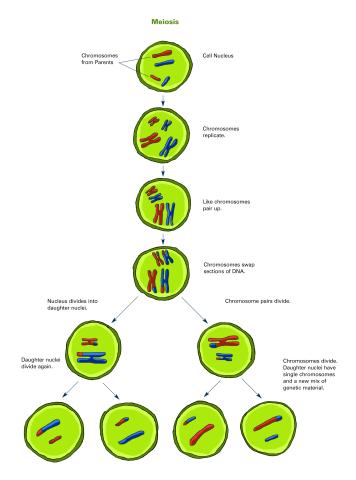
2546: Meiosis illustration (with labels)
2546: Meiosis illustration (with labels)
Meiosis is the process whereby a cell reduces its chromosomes from diploid to haploid in creating eggs or sperm. See image 2545 for an unlabeled version of this illustration. See image 2544 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
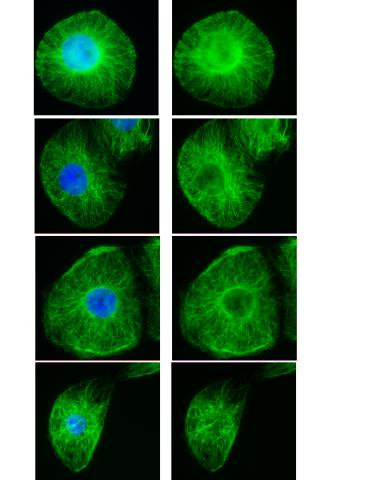
3443: Interphase in Xenopus frog cells
3443: Interphase in Xenopus frog cells
These images show frog cells in interphase. The cells are Xenopus XL177 cells, which are derived from tadpole epithelial cells. The microtubules are green and the chromosomes are blue. Related to 3442.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison.
View Media

2749: Cytoscape network wiring diagram 2
2749: Cytoscape network wiring diagram 2
This image integrates the thousands of known molecular and genetic interactions happening inside our bodies using a computer program called Cytoscape. Images like this are known as network wiring diagrams, but Cytoscape creator Trey Ideker somewhat jokingly calls them "hairballs" because they can be so complicated, intricate and hard to tease apart. Cytoscape comes with tools to help scientists study specific interactions, such as differences between species or between sick and diseased cells. Related to 2737.
Trey Ideker, University of California, San Diego
View Media
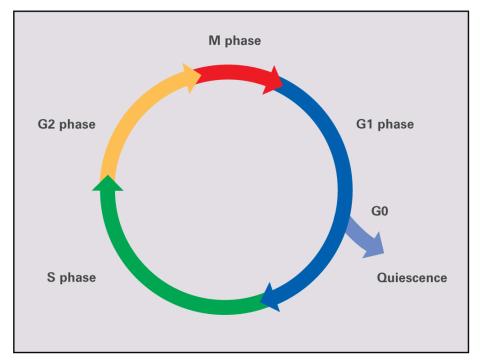
2499: Cell cycle (with labels)
2499: Cell cycle (with labels)
Cells progress through a cycle that consists of phases for growth (G1, S, and G2) and division (M). Cells become quiescent when they exit this cycle (G0). See image 2498 for an unlabeled version of this illustration.
Crabtree + Company
View Media
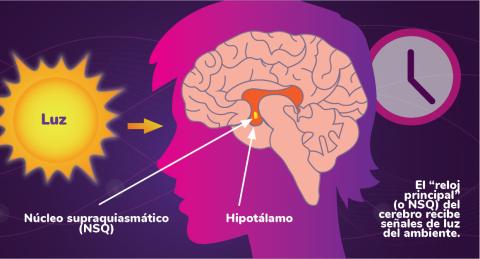
6614: Los ritmos circadianos y el núcleo supraquiasmático
6614: Los ritmos circadianos y el núcleo supraquiasmático
Los ritmos circadianos son cambios físicos, mentales y de comportamiento que siguen un ciclo de 24 horas. Los ritmos circadianos se ven influenciados por la luz y están regulados por el núcleo supraquiasmático del cerebro, a veces denominado el reloj principal.
Vea 6613 para la versión en inglés de esta infografía.
Vea 6613 para la versión en inglés de esta infografía.
NIGMS
View Media

2747: Cell division with late aligning chromosomes
2747: Cell division with late aligning chromosomes
This video shows an instance of abnormal mitosis where chromosomes are late to align. The video demonstrates the spindle checkpoint in action: just one unaligned chromosome can delay anaphase and the completion of mitosis. The cells shown are S3 tissue cultured cells from Xenopus laevis, African clawed frog.
Gary Gorbsky, Oklahoma Medical Research Foundation
View Media
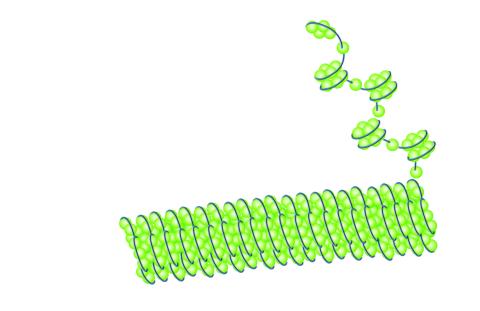
2560: Histones in chromatin
2560: Histones in chromatin
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2561 for a labeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media
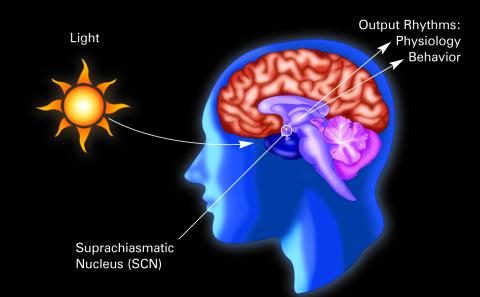
2569: Circadian rhythm (with labels)
2569: Circadian rhythm (with labels)
The human body keeps time with a master clock called the suprachiasmatic nucleus or SCN. Situated inside the brain, it's a tiny sliver of tissue about the size of a grain of rice, located behind the eyes. It sits quite close to the optic nerve, which controls vision, and this means that the SCN "clock" can keep track of day and night. The SCN helps control sleep and maintains our circadian rhythm--the regular, 24-hour (or so) cycle of ups and downs in our bodily processes such as hormone levels, blood pressure, and sleepiness. The SCN regulates our circadian rhythm by coordinating the actions of billions of miniature "clocks" throughout the body. These aren't actually clocks, but rather are ensembles of genes inside clusters of cells that switch on and off in a regular, 24-hour (or so) cycle in our physiological day.
Crabtree + Company
View Media
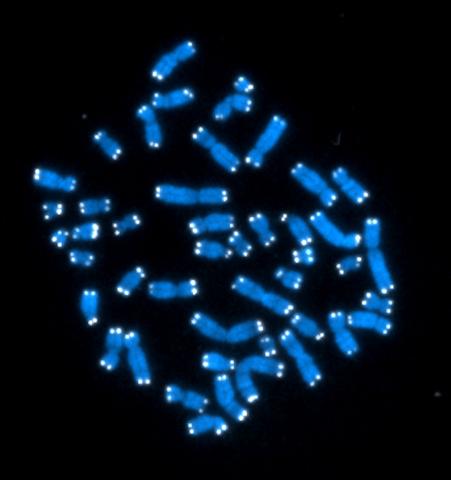
2626: Telomeres
2626: Telomeres
The 46 human chromosomes are shown in blue, with the telomeres appearing as white pinpoints. The DNA has already been copied, so each chromosome is actually made up of two identical lengths of DNA, each with its own two telomeres.
Hesed Padilla-Nash and Thomas Ried, the National Cancer Institute, a part of NIH
View Media
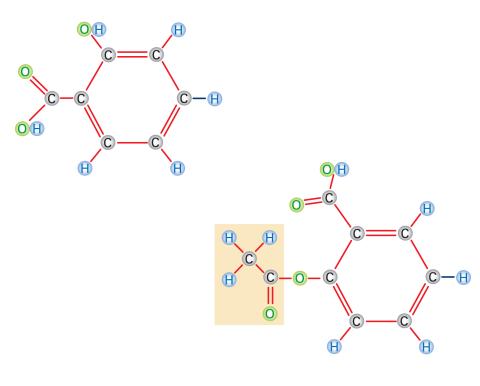
2529: Aspirin
2529: Aspirin
Acetylsalicylate (bottom) is the aspirin of today. Adding a chemical tag called an acetyl group (shaded box, bottom) to a molecule derived from willow bark (salicylate, top) makes the molecule less acidic (and easier on the lining of the digestive tract), but still effective at relieving pain. See image 2530 for a labeled version of this illustration. Featured in Medicines By Design.
Crabtree + Company
View Media
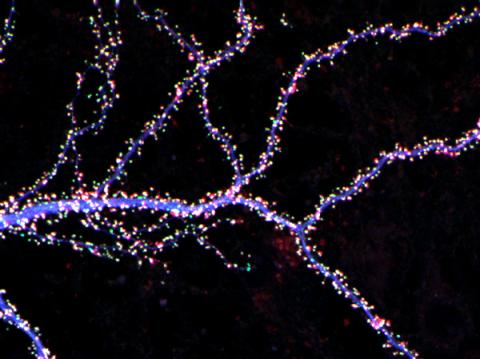
3686: Hippocampal neuron from rodent brain
3686: Hippocampal neuron from rodent brain
Hippocampal neuron from rodent brain with dendrites shown in blue. The hundreds of tiny magenta, green and white dots are the dendritic spines of excitatory synapses.
Shelley Halpain, UC San Diego
View Media

2319: Mapping metabolic activity
2319: Mapping metabolic activity
Like a map showing heavily traveled roads, this mathematical model of metabolic activity inside an E. coli cell shows the busiest pathway in white. Reaction pathways used less frequently by the cell are marked in red (moderate activity) and green (even less activity). Visualizations like this one may help scientists identify drug targets that block key metabolic pathways in bacteria.
Albert-László Barabási, University of Notre Dame
View Media
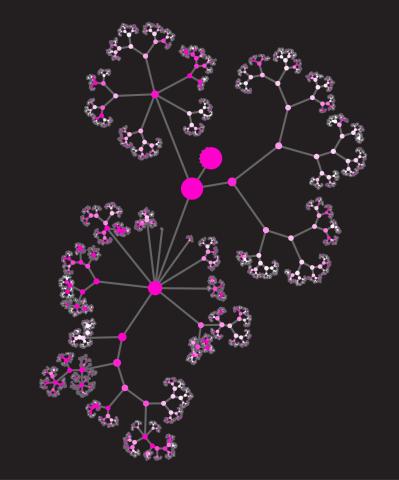
3436: Network diagram of genes, cellular components and processes (unlabeled)
3436: Network diagram of genes, cellular components and processes (unlabeled)
This image shows the hierarchical ontology of genes, cellular components and processes derived from large genomic datasets. From Dutkowski et al. A gene ontology inferred from molecular networks Nat Biotechnol. 2013 Jan;31(1):38-45. Related to 3437.
Janusz Dutkowski and Trey Ideker
View Media
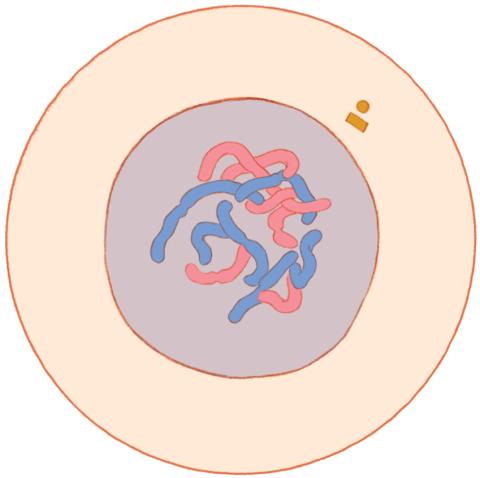
1316: Mitosis - interphase
1316: Mitosis - interphase
A cell in interphase, at the start of mitosis: Chromosomes duplicate, and the copies remain attached to each other. Mitosis is responsible for growth and development, as well as for replacing injured or worn out cells throughout the body. For simplicity, mitosis is illustrated here with only six chromosomes.
Judith Stoffer
View Media
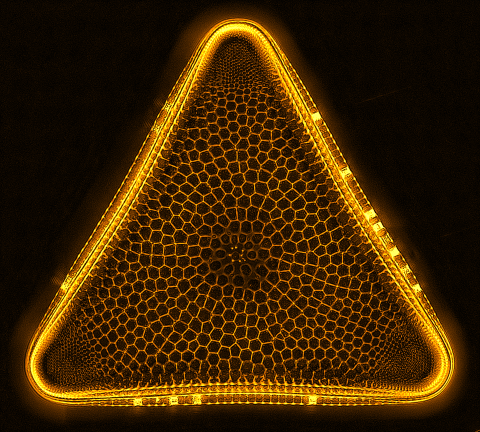
6962: Trigonium diatom
6962: Trigonium diatom
A Trigonium diatom imaged by a quantitative orientation-independent differential interference contrast (OI-DIC) microscope. Diatoms are single-celled photosynthetic algae with mineralized cell walls that contain silica and provide protection and support. These organisms form an important part of the plankton at the base of the marine and freshwater food chains. The width of this image is 90 μm.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
More information about the microscopy that produced this image can be found in the Journal of Microscopy paper “An Orientation-Independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model” by Malamy and Shribak.
Michael Shribak, Marine Biological Laboratory/University of Chicago.
View Media
